3KRY
 
 | | Crystal structure of MMP-13 in complex with SC-78080 | | Descriptor: | 1-(2-methoxyethyl)-N-oxo-4-({4-[4-(trifluoromethoxy)phenoxy]phenyl}sulfonyl)piperidine-4-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Kiefer, J.R, Williams, J.M, Becker, D.P. | | Deposit date: | 2009-11-19 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orally-active MMP-1 sparing alpha-tetrahydropyranyl and alpha-piperidinyl sulfone matrix metalloproteinase (MMP) inhibitors with efficacy in cancer, arthritis, and cardiovascular disease
J.Med.Chem., 53, 2010
|
|
5GXO
 
 | | Discovery of a compound that activates SIRT3 to deacetylate Manganese Superoxide Dismutase | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Lu, J, Li, J, Wu, M, Wang, J, Xia, Q. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A small molecule activator of SIRT3 promotes deacetylation and activation of manganese superoxide dismutase.
Free Radic. Biol. Med., 112, 2017
|
|
3LB9
 
 | | Crystal structure of the B. circulans cpA123 circular permutant | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | D'Angelo, I, Reitinger, S, Ludwiczek, M, Strynadka, N, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Circular permutation of Bacillus circulans xylanase: a kinetic and structural study.
Biochemistry, 49, 2010
|
|
8XUS
 
 | | JN.1 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4Z
 
 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X56
 
 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHZ
 
 | | BA.2.86 RBD in complex with hACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X55
 
 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHU
 
 | | Spike Trimer of BA.2.86 in complex with two hACE2s | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5R
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHW
 
 | | Spike Trimer of BA.2.86 with single RBD up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUR
 
 | | BA.2.86 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUU
 
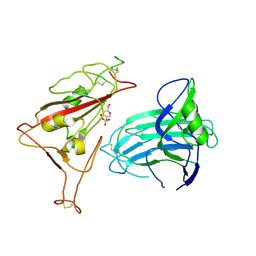 | | BA.2.86-T356K Spike Trimer in complex with heparan sulfate (Local refinement) | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X50
 
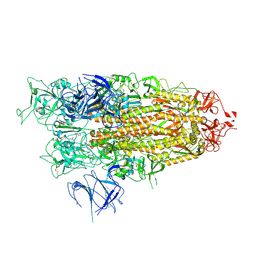 | | BA.2.86 Spike Trimer with ins483V mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4H
 
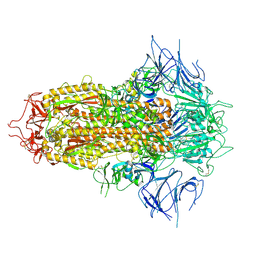 | | SARS-CoV-2 JN.1 Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5Q
 
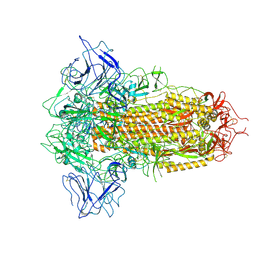 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUT
 
 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P, Mao, X. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8WHV
 
 | | Spike Trimer of BA.2.86 with three RBDs down | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
3ME3
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-{[4-(2,3-dihydro-1,4-benzodioxin-6-ylsulfonyl)-1,4-diazepan-1-yl]sulfonyl}aniline, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
8WHS
 
 | | Spike Trimer of BA.2.86 in complex with one hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-09-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
5LMZ
 
 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
4NXF
 
 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXB
 
 | | Crystal structure of iLOV-I486(2LT) at pH 7.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Li, J, Liu, X. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.561 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4NXE
 
 | | Crystal structure of iLOV-I486(2LT) at pH 6.5 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
4N9F
 
 | | Crystal structure of the Vif-CBFbeta-CUL5-ElOB-ElOC pentameric complex | | Descriptor: | Core-binding factor subunit beta, Cullin-5, Transcription elongation factor B polypeptide 1, ... | | Authors: | Guo, Y.Y, Dong, L.Y, Huang, Z.W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for hijacking CBF-b and CUL5 E3 ligase complex by HIV-1 Vif
Nature, 505, 2014
|
|
