7WFF
 
 | | Subcomplexes B,M and L in the Cylic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFD
 
 | | Left PSI in the cyclic electron transport supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Pan, X, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFE
 
 | | Right PSI in the cyclic electron transfer supercomplex NDH-PSI from Arabidopsis | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7WFG
 
 | | Subcomplexes A and E in NDH complex from Arabidopsis | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase subunit H, chloroplastic, ... | | Authors: | Pan, X.W, Li, M. | | Deposit date: | 2021-12-26 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.33 Å) | | Cite: | Supramolecular assembly of chloroplast NADH dehydrogenase-like complex with photosystem I from Arabidopsis thaliana.
Mol Plant, 15, 2022
|
|
7C9V
 
 | | E30 F-particle in complex with FcRn | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, MYRISTIC ACID, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9W
 
 | | E30 F-particle in complex with CD55 | | Descriptor: | Complement decay-accelerating factor, MYRISTIC ACID, SPHINGOSINE, ... | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9T
 
 | | Echovirus 30 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7C9U
 
 | | Echovirus 30 E-particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, K, Zhu, L, Sun, Y, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7CXN
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7DJJ
 
 | | Structure of four truncated and mutated forms of quenching protein lumenal domains | | Descriptor: | Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, SODIUM ION, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69806433 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJM
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, Protein SUPPRESSOR OF QUENCHING 1, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.70000112 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJL
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.96077824 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7DJK
 
 | | Structure of four truncated and mutated forms of quenching protein | | Descriptor: | CHLORIDE ION, Protein SUPPRESSOR OF QUENCHING 1, chloroplastic, ... | | Authors: | Yu, G.M, Pan, X.W, Li, M. | | Deposit date: | 2020-11-20 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.80145121 Å) | | Cite: | Structure of Arabidopsis SOQ1 lumenal region unveils C-terminal domain essential for negative regulation of photoprotective qH.
Nat.Plants, 8, 2022
|
|
7CXM
 
 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
7C9S
 
 | | Echovirus 30 F-particle | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Wang, K, Sun, Y, Zhu, L, Li, M, Zhao, X, Cui, L, Zhang, L, Gao, G, Zhai, W, Zhu, F, Rao, Z, Wang, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of Echovirus 30 in complex with its receptors inform a rational prediction for enterovirus receptor usage.
Nat Commun, 11, 2020
|
|
7YIM
 
 | | Cryo-EM structure of human Alpha-fetoprotein | | Descriptor: | Alpha-fetoprotein | | Authors: | Liu, N, Liu, K, Wu, C, Liu, Z, Li, M, Wang, J, Wang, H.W. | | Deposit date: | 2022-07-17 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Uniform thin ice on ultraflat graphene for high-resolution cryo-EM.
Nat.Methods, 20, 2023
|
|
7RW7
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 9 | | Descriptor: | (3'R)-2-[(cyclopropylmethyl)amino]-6-(4-methoxyphenyl)-1'-[(1H-pyrazol-5-yl)methyl]-5,6-dihydro-7H-spiro[pyrido[4,3-d]pyrimidine-8,3'-pyrrolidin]-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWH
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-41998 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-bromophenyl)-6-(4-methoxyphenyl)-2-[2,2,2-tris(fluoranyl)ethylamino]pyrido[4,3-d]pyrimidin-7-ol, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWG
 
 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW5
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | Descriptor: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
6AH4
 
 | | Structure of human P2X3 receptor in complex with ATP and Ca2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
6M1K
 
 | | USP7 in complex with a novel inhibitor | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7, methyl 4-[[4-[[3-[4-(aminomethyl)phenyl]-2-methyl-7-oxidanylidene-pyrazolo[4,3-d]pyrimidin-6-yl]methyl]-4-oxidanyl-piperidin-1-yl]methyl]-3-chloranyl-benzoate | | Authors: | Liu, S.J, Zhou, X.Y, Li, M.L, Sun, H.B, Wen, X.A. | | Deposit date: | 2020-02-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | N-benzylpiperidinol derivatives as novel USP7 inhibitors: Structure-activity relationships and X-ray crystallographic studies.
Eur.J.Med.Chem., 199, 2020
|
|
6LN0
 
 | |
3V4K
 
 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | CHLORIDE ION, DNA dC->dU-editing enzyme APOBEC-3G, MAGNESIUM ION, ... | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
3V4J
 
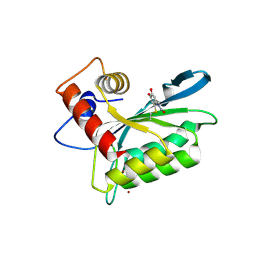 | | First-In-Class Small Molecule Inhibitors of the Single-strand DNA Cytosine Deaminase APOBEC3G | | Descriptor: | 4-[methyl(nitroso)amino]benzene-1,2-diol, DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Shandilya, S.M.D, Ali, A, Schiffer, C.A. | | Deposit date: | 2011-12-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | First-In-Class Small Molecule Inhibitors of the Single-Strand DNA Cytosine Deaminase APOBEC3G.
Acs Chem.Biol., 7, 2012
|
|
