5W4V
 
 | | Structure of RORgt bound to a tertiary alcohol | | Descriptor: | (R)-(4-chloro-2-methoxy-3-{[4-(1H-pyrazol-1-yl)phenyl]methyl}quinolin-6-yl)(1-methyl-1H-imidazol-5-yl)[6-(trifluoromethyl)pyridin-3-yl]methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Hars, U. | | Deposit date: | 2017-06-13 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 6-Substituted quinolines as ROR gamma t inverse agonists.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6TNS
 
 | | PI3K delta in complex with 2methoxyN[2methoxy5(7{[(2R)4(oxetan3 yl)morpholin2yl]methoxy}1,3dihydro2 benzofuran5yl)pyridin3yl]ethane1 sulfonamide | | Descriptor: | 2-methoxy-~{N}-[2-methoxy-5-[7-[[(2~{R})-4-(oxetan-3-yl)morpholin-2-yl]methoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]ethanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
6TNR
 
 | | PI3K delta in complex with N[5(7{2[4(2hydroxypropan2yl)piperidin1 yl]ethoxy}1,3dihydro2benzofuran5yl)2 methoxypyridin3yl]methanesulfonamide | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[2-methoxy-5-[7-[2-[4-(2-oxidanylpropan-2-yl)piperidin-1-yl]ethoxy]-1,3-dihydro-2-benzofuran-5-yl]pyridin-3-yl]methanesulfonamide | | Authors: | Convery, M.A, Rowland, P, Henley, Z.A, Barton, N, Down, K. | | Deposit date: | 2019-12-10 | | Release date: | 2020-01-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimization of Orally Bioavailable PI3K delta Inhibitors and Identification of Vps34 as a Key Selectivity Target.
J.Med.Chem., 63, 2020
|
|
6SWO
 
 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 2 | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
6VJT
 
 | |
6SWN
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH iBET-BD1 (GSK778) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(methoxymethyl)-1-[(1~{R})-1-phenylethyl]-8-[[(3~{S})-pyrrolidin-3-yl]methoxy]imidazo[4,5-c]quinolin-7-yl]-3,5-dimethyl-1,2-oxazole, Bromodomain-containing protein 4, ... | | Authors: | Chung, C. | | Deposit date: | 2019-09-22 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation.
Science, 368, 2020
|
|
7PI4
 
 | | FAK Protac GSK215 in complex with FAK and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-4-hydroxy-1-((S)-2-(2-(4-(3-methoxy-4-((4-((2-(methylcarbamoyl)phenyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)phenyl)piperazin-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Chung, C. | | Deposit date: | 2021-08-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery and Characterisation of Highly Cooperative FAK-Degrading PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6G9V
 
 | | Crystal structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase(AfUAP1) in complex with UDPGlcNAc, pyrophosphate and Mg2+ | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, UDP-N-acetylglucosamine pyrophosphorylase, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
6G9W
 
 | | Crystal Structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase in complex with UTP | | Descriptor: | UDP-N-acetylglucosamine pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
7LSZ
 
 | | Hsp90a N-terminal inhibitor | | Descriptor: | Heat shock protein HSP 90-alpha, {3-[(2,3-dihydro-1,4-benzodioxin-6-yl)sulfanyl]-4-hydroxyphenyl}(1,3-dihydro-2H-isoindol-2-yl)methanone | | Authors: | Balch, M, Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LT0
 
 | | Hsp90a N-terminal inhibitor | | Descriptor: | (1,3-dihydro-2H-isoindol-2-yl){3-[(3,4-dimethylphenyl)sulfanyl]-4-hydroxyphenyl}methanone, Heat shock protein HSP 90-alpha | | Authors: | Balch, M, Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-02-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Selective Inhibition of the Hsp90 alpha Isoform.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7QAR
 
 | | Serial crystallography structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-methyl uric acid at room temperature | | Descriptor: | (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, Uricase | | Authors: | Bui, S, Catapano, L, Zielinski, K, Yefanov, O, Murshudov, G.N, Oberthuer, D, Steiner, R.A. | | Deposit date: | 2021-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
2CAS
 
 | |
8C8T
 
 | | cryo-EM structure of BG505 SOSIP.664 HIV-1 Env trimer in complex with bNAbs EPTC112 and 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Baquero, E, Molinos-Albert, L, Mouquet, H. | | Deposit date: | 2023-01-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Anti-V1/V3-glycan broadly HIV-1 neutralizing antibodies in a post-treatment controller.
Cell Host Microbe, 31, 2023
|
|
4DPV
 
 | | PARVOVIRUS/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Chapman, M.S, Rossmann, M.G. | | Deposit date: | 1996-02-01 | | Release date: | 1997-04-01 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
4IR4
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone (GSK2834113A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-(morpholin-4-yl)-1-(pyridin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4IR5
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone (GSK2847449A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(hydroxymethyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4IR3
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-[7-amino-1-(pyrimidin-2-yl)indolizin-3-yl]ethanone (GSK2833282A) | | Descriptor: | 1,2-ETHANEDIOL, 1-[7-amino-1-(pyrimidin-2-yl)indolizin-3-yl]ethanone, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
4IR6
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with 1-{1-[2-(METHYLSULFONYL)PHENYL]-7-PHENOXYINDOLIZIN-3-YL}ETHANONE (GSK2838097A) | | Descriptor: | 1,2-ETHANEDIOL, 1-{1-[2-(methylsulfonyl)phenyl]-7-phenoxyindolizin-3-yl}ethanone, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Chaikuad, A, Felletar, I, Chung, C.W, Drewry, D, Chen, P, Filippakopoulos, P, Fedorov, O, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Characterization of GSK2801, a Selective Chemical Probe for the Bromodomains BAZ2A and BAZ2B.
J.Med.Chem., 59, 2016
|
|
5KJ9
 
 | |
3L00
 
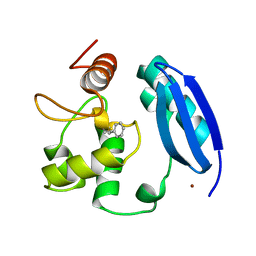 | |
3KZZ
 
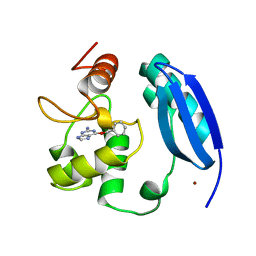 | |
4QL1
 
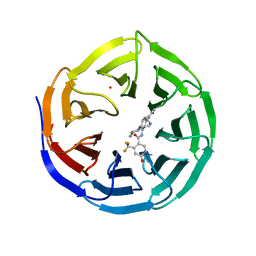 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
5TSZ
 
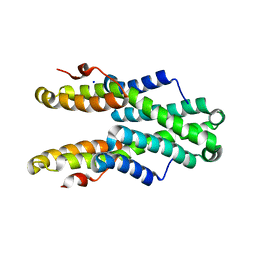 | | Crystal structure of Plasmodium vivax CelTOS | | Descriptor: | Pv cell-traversal protein, SODIUM ION | | Authors: | Tolia, N.H, Jimah, J.R. | | Deposit date: | 2016-10-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Malaria parasite CelTOS targets the inner leaflet of cell membranes for pore-dependent disruption.
Elife, 5, 2016
|
|
5UFR
 
 | | Structure of RORgt bound to | | Descriptor: | (S)-[4-chloro-2-(dimethylamino)-3-phenylquinolin-6-yl](1-methyl-1H-imidazol-5-yl)(pyridin-4-yl)methanol, Nuclear receptor ROR-gamma | | Authors: | Spurlino, J, Abad, M. | | Deposit date: | 2017-01-05 | | Release date: | 2017-04-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Identification and structure activity relationships of quinoline tertiary alcohol modulators of ROR gamma t.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
