3O0I
 
 | |
3O2F
 
 | |
4XU6
 
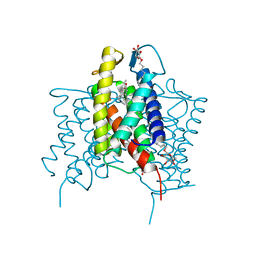 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
6H6V
 
 | |
6H6X
 
 | | Structure of an evolved dimeric form of the UbiD-class enzyme HmfF from Pelotomaculum thermopropionicum in complex with prFMN | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3-polyprenyl-4-hydroxybenzoate decarboxylase and related decarboxylases, CALCIUM ION, ... | | Authors: | Payne, K.A.P, Leys, D. | | Deposit date: | 2018-07-30 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Enzymatic Carboxylation of 2-Furoic Acid Yields 2,5-Furandicarboxylic Acid (FDCA).
Acs Catalysis, 9, 2019
|
|
2RHS
 
 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
2RHQ
 
 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
5MPS
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
3NEG
 
 | | Pyrabactin-bound PYL1 structure in the open and close forms | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1, BROMIDE ION | | Authors: | Yan, N. | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Functional mechanism of the ABA agonist pyrabactin
J.Biol.Chem., 285, 2010
|
|
3NEF
 
 | | High-resolution pyrabactin-bound PYL1 structure | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL1 | | Authors: | Yan, N. | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional mechanism of the ABA agonist pyrabactin
J.Biol.Chem., 285, 2010
|
|
6BYO
 
 | |
3NR4
 
 | | Pyrabactin-bound PYL2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Yan, N. | | Deposit date: | 2010-06-30 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Single amino acid alteration between Valine and Isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2
J.Biol.Chem., 285, 2010
|
|
2VQD
 
 | |
2VPQ
 
 | |
2VR1
 
 | |
8WJQ
 
 | | Cryo-EM structure of URAT1(R477S)-Urate complex | | Descriptor: | Solute carrier family 22 member 12, URIC ACID | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8WJH
 
 | | Cryo-EM structure of OAT4 | | Descriptor: | CHLORIDE ION, Solute carrier family 22 member 11 | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8WJG
 
 | | Cryo-EM structure of URAT1(R477S) | | Descriptor: | Solute carrier family 22 member 12 | | Authors: | Qian, H.W, He, J.J. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the transport and substrate selection of human urate transporter 1.
Cell Rep, 43, 2024
|
|
8J2X
 
 | | Saccharothrix syringae photocobilins protein, light state | | Descriptor: | BILIVERDINE IX ALPHA, COBALAMIN, Cobalamin-binding protein, ... | | Authors: | Zhang, S, Poddar, H, Levy, C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
7MCE
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol | | Descriptor: | 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol, Bromodomain-containing protein 4 | | Authors: | Sheriff, S. | | Deposit date: | 2021-04-02 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
8J2Y
 
 | | Acidimicrobiaceae bacterium photocobilins protein, dark state | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, S, Poddar, H, Levy, W.C, Leys, D. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Photocobilins integrate B12 and bilin photochemistry for enzyme control.
Nat Commun, 15, 2024
|
|
4GBY
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose | | Descriptor: | D-xylose-proton symporter, beta-D-xylopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GBZ
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose | | Descriptor: | D-xylose-proton symporter, beta-D-glucopyranose, nonyl beta-D-glucopyranoside | | Authors: | Sun, L.F, Zeng, X, Yan, C.Y, Yan, N. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
4GC0
 
 | | The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose | | Descriptor: | 6-bromo-6-deoxy-beta-D-glucopyranose, D-xylose-proton symporter, nonyl beta-D-glucopyranoside | | Authors: | Yan, N, Sun, L.F, Zeng, X, Yan, C.Y. | | Deposit date: | 2012-07-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a bacterial homologue of glucose transporters GLUT1-4.
Nature, 490, 2012
|
|
