4PII
 
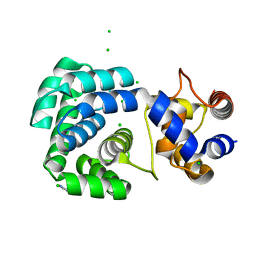 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4BPM
 
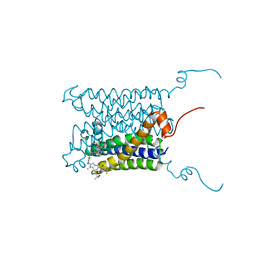 | | Crystal structure of a human integral membrane enzyme | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Li, D, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
Cryst.Growth Des., 14, 2014
|
|
6EQP
 
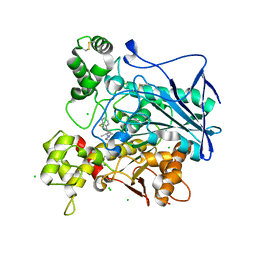 | | Human butyrylcholinesterase in complex with ethopropazine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Rosenberry, T.L, Macdonald, I.R, Darvesh, S. | | Deposit date: | 2017-10-14 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.349417 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
6JVY
 
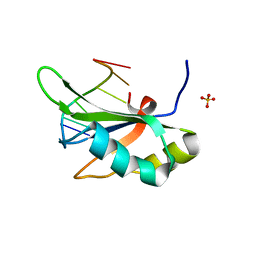 | | Crystal structure of RBM38 in complex with single-stranded DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*GP*TP*GP*TP*GP*TP*GP*TP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
6EP4
 
 | | Human butyrylcholinesterase in complex with decamethonium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Rosenberry, T.L, Macdonald, I.R, Darvesh, S. | | Deposit date: | 2017-10-10 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.30005383 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
6EQQ
 
 | | Human butyrylcholinesterase in complex with huprine 19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Nachon, F, Brazzolotto, X, Wandhammer, M, Trovaslet-Leroy, M, Rosenberry, T.L, Macdonald, I.R, Darvesh, S. | | Deposit date: | 2017-10-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.40000439 Å) | | Cite: | Comparison of the Binding of Reversible Inhibitors to Human Butyrylcholinesterase and Acetylcholinesterase: A Crystallographic, Kinetic and Calorimetric Study.
Molecules, 22, 2017
|
|
3EAD
 
 | | Crystal structure of CALX-CBD1 | | Descriptor: | CALCIUM ION, GLYCEROL, Na/Ca exchange protein | | Authors: | Zheng, L, Wang, M. | | Deposit date: | 2008-08-25 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
4CJC
 
 | | orthorhombic crystal form of Bogt6a E192Q in complex with UDP-GalNAc, UDP, GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSYLTRANSFERASE FAMILY 6, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products.
J.Biol.Chem., 289, 2014
|
|
4CJB
 
 | | orthorhombic crystal form of Bogt6a E192Q in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSYLTRANSFERASE FAMILY 6 | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products.
J.Biol.Chem., 289, 2014
|
|
2AKJ
 
 | | Structure of spinach nitrite reductase | | Descriptor: | Ferredoxin--nitrite reductase, chloroplast, IRON/SULFUR CLUSTER, ... | | Authors: | Swamy, U, Wang, M, Tripathy, J.N, Kim, S.-K, Hirasawa, M, Knaff, D.B, Allen, J.P. | | Deposit date: | 2005-08-03 | | Release date: | 2006-01-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Spinach Nitrite Reductase: Implications for Multi-electron Reactions by the Iron-Sulfur:Siroheme Cofactor
Biochemistry, 44, 2005
|
|
4CJ8
 
 | | monoclinic crystal form of Bogt6a E192Q in complex with UDP-GalNAc, UDP and GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, GLYCEROL, ... | | Authors: | Pham, T, Stinson, B, Thiyagarajan, N, Lizotte-Waniewski, M, Brew, K, Acharya, K.R. | | Deposit date: | 2013-12-19 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Complexes of a Metal-Independent Glycosyltransferase Gt6 from Bacteroides Ovatus with Udp-Galnac and its Hydrolysis Products
J.Biol.Chem., 289, 2014
|
|
4H75
 
 | | Crystal structure of human Spindlin1 in complex with a histone H3K4(me3) peptide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Histone H3, ... | | Authors: | Yang, N, Wang, W, Wang, Y, Wang, M, Zhao, Q, Rao, Z, Zhu, B, Xu, R.M. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2P05
 
 | | Structural Insights into the Evolution of a Non-Biological Protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Smith, M, Rosenow, M, Wang, M, Allen, J.P, Szostak, J.W, Chaput, J.C. | | Deposit date: | 2007-02-28 | | Release date: | 2007-06-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the evolution of a non-biological protein: importance of surface residues in protein fold optimization.
PLoS ONE, 2, 2007
|
|
6LW5
 
 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
5CD7
 
 | | Crystal structure of the NTD L199M of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5N6L
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt C387A mutant from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
5CD8
 
 | | Crystal structure of the NTD of Drosophila Oskar protein | | Descriptor: | GLYCEROL, Maternal effect protein oskar | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CD9
 
 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5N6H
 
 | | Structure of the membrane integral lipoprotein N-acyltransferase Lnt from E. coli | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Apolipoprotein N-acyltransferase, GLYCEROL | | Authors: | Huang, C.-Y, Boland, C, Howe, N, Wiktor, M, Vogeley, L, Weichert, D, Bailey, J, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of the membrane integral N-acyltransferase step in bacterial lipoprotein synthesis.
Nat Commun, 8, 2017
|
|
7QN3
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 13) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QLU
 
 | | Endothiapepsin apo at 100K | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, Endothiapepsin | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QLT
 
 | | Endothiapepsin in complex with ligand TL00150 in 10% DMSO at 100K | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QLY
 
 | | Endothiapepsin apo at 298K | | Descriptor: | Endothiapepsin | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QMZ
 
 | | Endothiapepsin in complex with compound TL00150 at room-temperature (temperature ramping up structure 9) | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QLZ
 
 | | Endothiapepsin in 5% DMSO at 298 K | | Descriptor: | Endothiapepsin | | Authors: | Huang, C.Y, Aumonier, S, Wang, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Probing ligand binding of endothiapepsin by `temperature-resolved' macromolecular crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
