1MX4
 
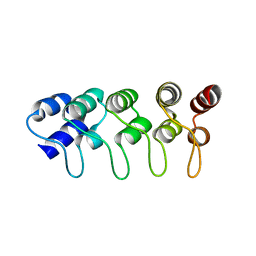 | | Structure of p18INK4c (F82Q) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1MUS
 
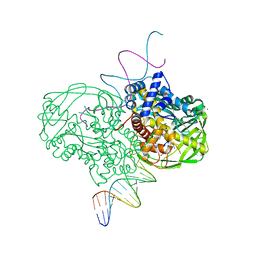 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
1MX6
 
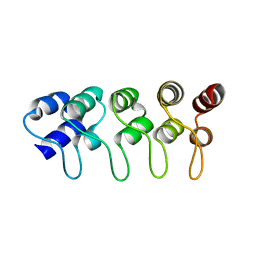 | | Structure of p18INK4c (F92N) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
1KAQ
 
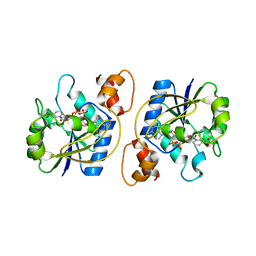 | | Structure of Bacillus subtilis Nicotinic Acid Mononucleotide Adenylyl Transferase | | Descriptor: | NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Olland, A.M, Underwood, K.W, Czerwinski, R.M, Lo, M.C, Aulabaugh, A, Bard, J, Stahl, M.L, Somers, W.S, Sullivan, F.X, Chopra, R. | | Deposit date: | 2001-11-02 | | Release date: | 2002-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase.
J.Biol.Chem., 277, 2002
|
|
1KV8
 
 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
5IHY
 
 | |
5ITG
 
 | |
5KNR
 
 | | E. coli HPRT in complexed with 9-[(N-phosphonoethyl-N-phosphonoethoxyethyl)-2-aminoethyl]-guanine | | Descriptor: | (2-{[2-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)ethyl][2-(2-phosphonoethoxy)ethyl]amino}ethyl)phosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.864 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNU
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with 9-[N,N-(Bis-3-phosphonopropyl)aminomethyl]-9-deazahypoxanthine | | Descriptor: | 3-[(4-oxidanylidene-3,5-dihydropyrrolo[3,2-d]pyrimidin-7-yl)methyl-(3-phosphonopropyl)amino]propylphosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Baszczynski, O, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNX
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with {[(2-[(Hypoxanthin-9H-yl)methyl]propane-1,3-diyl)bis(oxy)]bis- (methylene)}diphosphonic Acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [2-[(6-oxidanylidene-1~{H}-purin-9-yl)methyl]-3-(phosphonomethoxy)propoxy]methylphosphonic acid | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNT
 
 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with 2-((2,3-Dihydroxypropyl)(2-(hypoxanthin-9-yl)ethyl)amino)ethylphosphonic acid | | Descriptor: | 2-[[(2~{S})-2,3-bis(oxidanyl)propyl]-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl]amino]ethylphosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNV
 
 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-Phosphonoethyl-N-phosphonobutyl)-2-aminoethyl]-hypoxanthine | | Descriptor: | 2-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl-(4-phosphonobutyl)amino]ethylphosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5KNQ
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with [3S,4R]-(4-(Guanin-9-yl)pyrrolidin-3-yl)oxymethanephosphonic acid and pyrophosphate | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
5KNY
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with (3-((3R,4R)-3-(Guanin-9-yl)-4-((S)-2-hydroxy-2-phosphonoethoxy)pyrrolidin-1-yl)-3-oxopropyl)phosphonic acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, [3-[(3~{R},4~{R})-3-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-[(2~{S})-2-oxidanyl-2-phosphono-ethoxy]pyrrolidin-1-y l]-3-oxidanylidene-propyl]phosphonic acid | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
1X78
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1, [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1XBY
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/T169A mutant with bound D-ribulose 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, MAGNESIUM ION, RIBULOSE-5-PHOSPHATE | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
1XFR
 
 | |
1X7B
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH ERB-041 | | Descriptor: | 2-(3-FLUORO-4-HYDROXYPHENYL)-7-VINYL-1,3-BENZOXAZOL-5-OL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1XBZ
 
 | | Crystal structure of 3-keto-L-gulonate 6-phosphate decarboxylase E112D/R139V/T169A mutant with bound L-xylulose 5-phosphate | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, L-XYLULOSE 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Akana, J, Gerlt, J.A, Rayment, I. | | Deposit date: | 2004-08-31 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: structural basis for catalytic promiscuity in wild-type and designed mutants of 3-keto-L-gulonate 6-phosphate decarboxylase
Biochemistry, 44, 2005
|
|
5XLJ
 
 | | Crystal structure of the flagellar cap protein flid D2-D3 domains from serratia marcescens in Space group P432 | | Descriptor: | CHLORIDE ION, Flagellar hook-associated protein 2, SODIUM ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
1ZGW
 
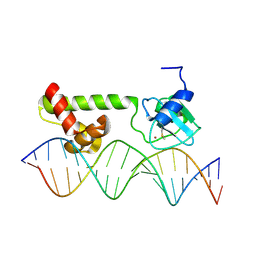 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1X76
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR BETA COMPLEXED WITH WAY-697 | | Descriptor: | 5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-CARBONITRILE, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
1X7E
 
 | | CRYSTAL STRUCTURE OF ESTROGEN RECEPTOR ALPHA COMPLEXED WITH WAY-244 | | Descriptor: | Estrogen receptor 1 (alpha), [5-HYDROXY-2-(4-HYDROXYPHENYL)-1-BENZOFURAN-7-YL]ACETONITRILE, steroid receptor coactivator-3 | | Authors: | Manas, E.S, Unwalla, R.J, Xu, Z.B, Malamas, M.S, Miller, C.P, Harris, H.A, Hsiao, C, Akopian, T, Hum, W.T, Malakian, K, Wolfrom, S, Bapat, A, Bhat, R.A, Stahl, M.L, Somers, W.S, Alvarez, J.C. | | Deposit date: | 2004-08-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Estrogen Receptor-Beta Selective Ligands
J.Am.Chem.Soc., 126, 2004
|
|
5YHH
 
 | | Crystal structure of YiiM from Geobacillus stearothermophilus | | Descriptor: | Uncharacterized conserved protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
5YHI
 
 | | Crystal structure of YiiM from Escherichia coli | | Descriptor: | PHOSPHATE ION, Protein YiiM | | Authors: | Namgung, B, Kim, J.H, Song, W.S, Yoon, S.I. | | Deposit date: | 2017-09-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of the hydroxylaminopurine resistance protein, YiiM, and its putative molybdenum cofactor-binding catalytic site.
Sci Rep, 8, 2018
|
|
