6CCR
 
 | | Selenomethionyl derivative of a GID4 fragment | | Descriptor: | Glucose-induced degradation protein 4 homolog, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CCU
 
 | | Complex between a GID4 fragment and a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Short peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CD9
 
 | | GID4 in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6BHH
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6C1Y
 
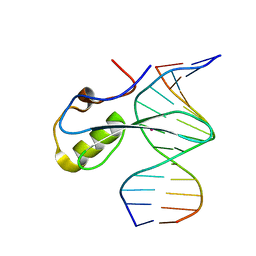 | | mbd of human mecp2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Bian, C, Tempel, W, Wernimont, A.K, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Plasticity at the DNA recognition site of the MeCP2 mCG-binding domain.
Biochim Biophys Acta Gene Regul Mech, 1862, 2019
|
|
6BY9
 
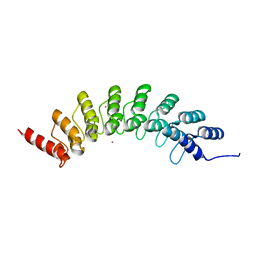 | | Crystal structure of EHMT1 | | Descriptor: | Histone-lysine N-methyltransferase EHMT1, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Wei, Y, Li, A, Tempel, W, Han, S, Sunnerhagen, M, Penn, L, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-20 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of EHMT1
to be published
|
|
6C2F
 
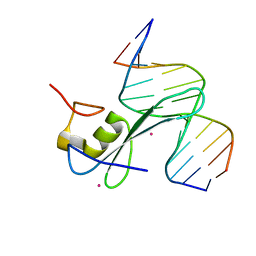 | | MBD2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-08 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MBD2 in complex with methylated DNA
to be published
|
|
6CDG
 
 | | GID4 fragment in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Hexapeptide PGLWKS, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6DUB
 
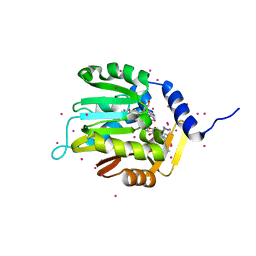 | | Crystal structure of a methyltransferase | | Descriptor: | Alpha N-terminal protein methyltransferase 1B, GLYCEROL, RCC1, ... | | Authors: | Dong, C, Tempel, W, Li, Y, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | An asparagine/glycine switch governs product specificity of human N-terminal methyltransferase NTMT2.
Commun Biol, 1, 2018
|
|
4E71
 
 | | Crystal structure of the RHO GTPASE binding domain of Plexin B2 | | Descriptor: | Plexin-B2, SODIUM ION | | Authors: | Guan, X, Wang, H, Tempel, W, Tong, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the RHO GTPASE binding domain of Plexin B2
to be published
|
|
4E74
 
 | | Crystal structure of the RHO GTPASE BINDING DOMAIN of Plexin A4A | | Descriptor: | Plexin-A4, UNKNOWN ATOM OR ION | | Authors: | Guan, X, Wang, H, Tempel, W, Dong, A, Tong, Y, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of the RHO GTPASE BINDING DOMAIN of Plexin A4A
to be published
|
|
4EDU
 
 | | The MBT repeats of human SCML2 in a complex with histone H2A peptide | | Descriptor: | Histone H2A.J peptide, Sex comb on midleg-like protein 2 | | Authors: | Nady, N, Amaya, M.F, Tempel, W, Ravichandran, M, Arrowsmith, C.H. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Histone recognition by human malignant brain tumor domains.
J.Mol.Biol., 423, 2012
|
|
4EZA
 
 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
4DRZ
 
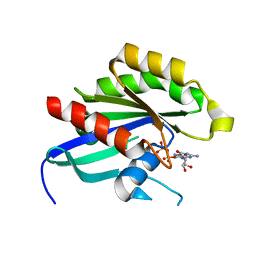 | | Crystal structure of human RAB14 | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, Ras-related protein Rab-14 | | Authors: | Wang, J, Tempel, W, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-17 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human RAB14
to be published
|
|
4FK7
 
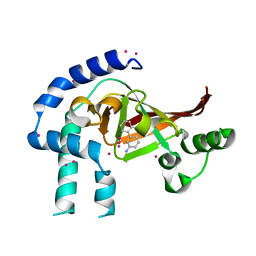 | | Crystal structure of Certhrax catalytic domain | | Descriptor: | CHLORIDE ION, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-12 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
4FMW
 
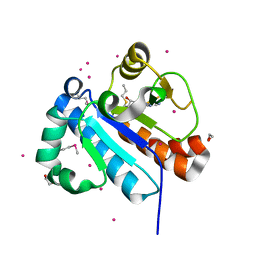 | | Crystal structure of methyltransferase domain of human RNA (guanine-9-) methyltransferase domain containing protein 2 | | Descriptor: | 1,2-ETHANEDIOL, POTASSIUM ION, RNA (guanine-9-)-methyltransferase domain-containing protein 2, ... | | Authors: | Dong, A, Zeng, H, Loppnau, P, Tempel, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of methyltransferase domain of human RNA (guanine-9-) methyltransferase domain containing protein 2
TO BE PUBLISHED
|
|
4FL6
 
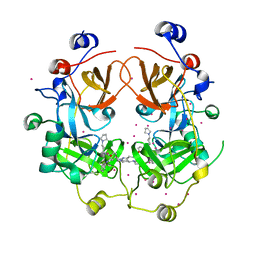 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
4FUK
 
 | | Aminopeptidase from Trypanosoma brucei | | Descriptor: | GLYCEROL, Methionine aminopeptidase, UNKNOWN ATOM OR ION, ... | | Authors: | El Bakkouri, M, Tempel, W, Osman, K.T, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lin, Y.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aminpeptidase from Trypanosoma brucei (CASP Target)
To be Published
|
|
4FO9
 
 | | Crystal structure of the E3 SUMO Ligase PIAS2 | | Descriptor: | E3 SUMO-protein ligase PIAS2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Hu, J, Dobrovetsky, E, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the E3 SUMO Ligase PIAS2
to be published
|
|
4GI7
 
 | | Crystal structure of Klebsiella pneumoniae pantothenate kinase in complex with a pantothenate analogue | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-{3-oxo-3-[(pyridin-3-ylmethyl)amino]propyl}butanamide, ADENOSINE-5'-DIPHOSPHATE, Pantothenate kinase, ... | | Authors: | Li, B, Tempel, W, Smil, D, Bolshan, Y, Hong, B.S, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-08 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Klebsiella pneumoniae pantothenate kinase in complex with N-substituted pantothenamides.
Proteins, 81, 2013
|
|
4GF1
 
 | | Crystal Structure of Certhrax | | Descriptor: | Putative ADP-ribosyltransferase Certhrax, UNKNOWN ATOM OR ION | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-02 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
4F7W
 
 | | Crystal structure of Klebsiella pneumoniae pantothenate kinase in complex with N-pentylpantothenamide | | Descriptor: | (2R)-2,4-dihydroxy-3,3-dimethyl-N-[3-oxo-3-(pentylamino)propyl]butanamide, ADENOSINE-5'-DIPHOSPHATE, Pantothenate kinase, ... | | Authors: | Li, B, Tempel, W, Smil, D, Bolshan, Y, Hong, B.S, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-16 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of Klebsiella pneumoniae pantothenate kinase in complex with N-substituted pantothenamides.
Proteins, 81, 2013
|
|
4FLB
 
 | | CID of human RPRD2 | | Descriptor: | PRASEODYMIUM ION, Regulation of nuclear pre-mRNA domain-containing protein 2, SULFATE ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4FLA
 
 | | Crystal structure of human RPRD1B, carboxy-terminal domain | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4FU3
 
 | | CID of human RPRD1B | | Descriptor: | CHLORIDE ION, Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CID of human RPRD1B
TO BE PUBLISHED
|
|
