3SZ9
 
 | |
3UPX
 
 | |
2R7E
 
 | | Crystal Structure Analysis of Coagulation Factor VIII | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Stoddard, B.L, Shen, B.W. | | Deposit date: | 2007-09-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The tertiary structure and domain organization of coagulation factor VIII.
Blood, 111, 2008
|
|
6A4V
 
 | | Open Reading frame 49 | | Descriptor: | 49 protein | | Authors: | Hwang, K.Y, Song, M.J, Kim, J.S, Cheong, W.C. | | Deposit date: | 2018-06-21 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based mechanism of action of a viral poly(ADP-ribose) polymerase 1-interacting protein facilitating virus replication.
Iucrj, 5, 2018
|
|
8IU0
 
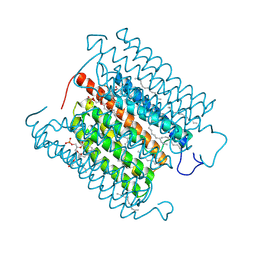 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 H225F mutant in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Nakamura, S, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2023-03-23 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
2VXP
 
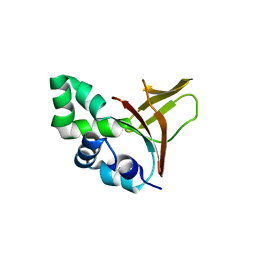 | | The fourth FAS1 domain structure of human Bigh3 | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA-INDUCED PROTEIN IG-H3 | | Authors: | Yoo, J.-H, Cho, H.-S. | | Deposit date: | 2008-07-08 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structural Analysis of the Fas1 Domain 4 of Big-H3 for Relationship with Corneal Dystrophy
To be Published
|
|
5SY1
 
 | | Structure of the STRA6 receptor for retinol uptake in complex with calmodulin | | Descriptor: | CALCIUM ION, CHOLESTEROL, Calmodulin, ... | | Authors: | Clarke, O.B, Chen, Y, Mancia, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the STRA6 receptor for retinol uptake.
Science, 353, 2016
|
|
8H86
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR1 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR1, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
8H87
 
 | | Cryo-EM structure of the potassium-selective channelrhodopsin HcKCR2 in lipid nanodisc | | Descriptor: | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, HcKCR2, PALMITIC ACID, ... | | Authors: | Tajima, S, Kim, Y, Yamashita, K, Fukuda, M, Deisseroth, K, Kato, H.E. | | Deposit date: | 2022-10-21 | | Release date: | 2023-09-06 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | Structural basis for ion selectivity in potassium-selective channelrhodopsins.
Cell, 186, 2023
|
|
3V51
 
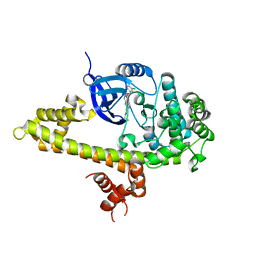 | |
3V5P
 
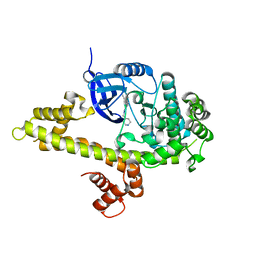 | |
4R1N
 
 | |
4Y5Q
 
 | |
4K86
 
 | |
4OWI
 
 | | peptide structure | | Descriptor: | p53LZ2 | | Authors: | Lee, J.-H. | | Deposit date: | 2014-02-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.202 Å) | | Cite: | Protein grafting of p53TAD onto a leucine zipper scaffold generates a potent HDM dual inhibitor.
Nat Commun, 5, 2014
|
|
4K87
 
 | | Crystal structure of human prolyl-tRNA synthetase (substrate bound form) | | Descriptor: | ADENOSINE, PROLINE, Proline--tRNA ligase, ... | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3P2F
 
 | | Crystal structure of TofI in an apo form | | Descriptor: | AHL synthase | | Authors: | Yu, S, Rhee, S. | | Deposit date: | 2010-10-02 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3P2H
 
 | |
4K88
 
 | | Crystal structure of human prolyl-tRNA synthetase (halofuginone bound form) | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, Proline--tRNA ligase, ZINC ION | | Authors: | Hwang, K.Y, Son, J.H, Lee, E.H. | | Deposit date: | 2013-04-18 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | Conformational changes in human prolyl-tRNA synthetase upon binding of the substrates proline and ATP and the inhibitor halofuginone.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5F3K
 
 | | X-Ray Crystallographic Structure of hTrap1 N-terminal Domain-apo | | Descriptor: | Heat shock protein 75 kDa, mitochondrial | | Authors: | Sung, N, Lee, J, Kim, J, Chang, C, Joachimiak, A, Lee, S, Tsai, F.T.F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mitochondrial Hsp90 is a ligand-activated molecular chaperone coupling ATP binding to dimer closure through a coiled-coil intermediate.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3RIG
 
 | |
2GJL
 
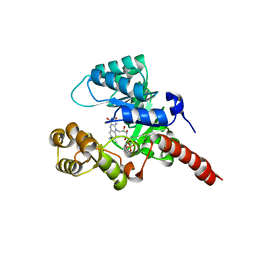 | | Crystal Structure of 2-nitropropane dioxygenase | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical protein PA1024 | | Authors: | Suh, S.W. | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of 2-Nitropropane Dioxygenase Complexed with FMN and Substrate: identification of the catalytic base
J.Biol.Chem., 281, 2006
|
|
3RIY
 
 | |
2GJN
 
 | |
8FBW
 
 | | Crystal structure of SIV-1 V2 antibody NCI05 in complex with a V2 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, T, Kwong, P.D, Van Wazer, D.J. | | Deposit date: | 2022-11-30 | | Release date: | 2023-01-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Effect of Passive Administration of Monoclonal Antibodies Recognizing Simian Immunodeficiency Virus (SIV) V2 in CH59-Like Coil/Helical or beta-Sheet Conformations on Time of SIV mac251 Acquisition.
J.Virol., 97, 2023
|
|
