7QVH
 
 | |
6M32
 
 | | Cryo-EM structure of FMO-RC complex from green sulfur bacteria | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2-[(1E,3E,5E,7E,9E,11E,13E,15E,17E,19E)-3,7,12,16,20,24-hexamethylpentacosa-1,3,5,7,9,11,13,15,17,19,23-undecaenyl]-1,3,4-trimethyl-benzene, ... | | Authors: | Chen, J.H, Zhang, X. | | Deposit date: | 2020-03-02 | | Release date: | 2020-11-25 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the photosynthetic complex from a green sulfur bacterium.
Science, 370, 2020
|
|
8JM9
 
 | |
8JMI
 
 | |
8JMA
 
 | |
8JME
 
 | |
8JMH
 
 | |
7XUK
 
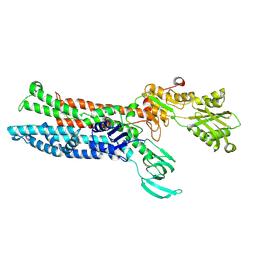 | |
7XUN
 
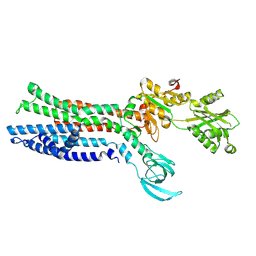 | |
6UKB
 
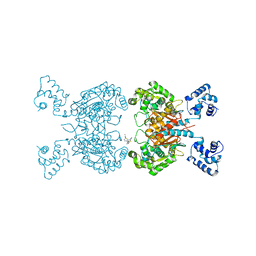 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-[5-(4-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]oxy}piperidin-1-yl)-1,3,4-thiadiazol-2-yl]acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00020
To Be Published
|
|
8GWR
 
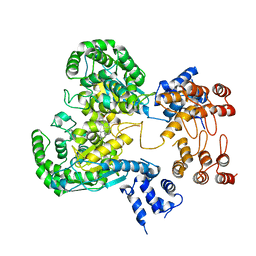 | | Near full length Kidney type Glutaminase in complex with 2,2-Dimethyl-2,3-Dihydrobenzo[a] Phenanthridin-4(1H)-one (DDP) | | Descriptor: | 2,2-dimethyl-1,3-dihydrobenzo[a]phenanthridin-4-one, Glutaminase kidney isoform, mitochondrial | | Authors: | Shankar, S, Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | A novel allosteric site employs a conserved inhibition mechanism in human kidney-type glutaminase.
Febs J., 290, 2023
|
|
2K0T
 
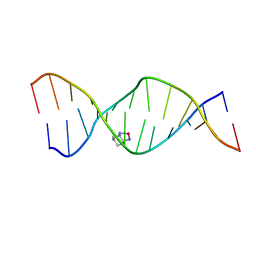 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-14 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
2K0U
 
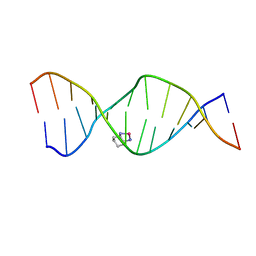 | | High Resolution Solution NMR Structures of Oxaliplatin-DNA Adduct | | Descriptor: | CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II), DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
2K0V
 
 | | High Resolution Solution NMR Structures of Undamaged DNA Dodecamer Duplex | | Descriptor: | DNA (5'-D(*DCP*DCP*DTP*DCP*DTP*DGP*DGP*DTP*DCP*DTP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DAP*DGP*DAP*DCP*DCP*DAP*DGP*DAP*DGP*DG)-3') | | Authors: | Bhattacharyya, D, King, C.L, Chaney, S.G, Campbell, S.L. | | Deposit date: | 2008-02-15 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Flanking Bases Influence the Nature of DNA Distortion by Platinum 1,2-Intrastrand (GG) Cross-Links.
Plos One, 6, 2011
|
|
8TBF
 
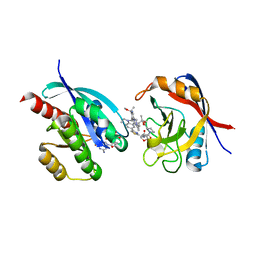 | | Tricomplex of RMC-7977, KRAS WT, and CypA | | Descriptor: | (1R,5S,6r)-N-[(1P,7S,9S,13S,20M)-20-{5-(4-cyclopropylpiperazin-1-yl)-2-[(1S)-1-methoxyethyl]pyridin-3-yl}-21-ethyl-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-3-oxabicyclo[3.1.0]hexane-6-carboxamide, 1,2-ETHANEDIOL, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Concurrent inhibition of oncogenic and wild-type RAS-GTP for cancer therapy.
Nature, 629, 2024
|
|
8JKM
 
 | | RN-1747 bound state of mTRPV4 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4 | | Authors: | Zhen, W.X, Yang, F. | | Deposit date: | 2023-06-01 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis of ligand activation and inhibition in a mammalian TRPV4 ion channel.
Cell Discov, 9, 2023
|
|
5C9C
 
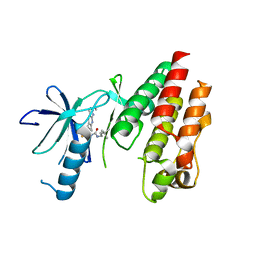 | | CRYSTAL STRUCTURE OF BRAF(V600E) IN COMPLEX WITH LY3009120 COMPND | | Descriptor: | 1-(3,3-dimethylbutyl)-3-{2-fluoro-4-methyl-5-[7-methyl-2-(methylamino)pyrido[2,3-d]pyrimidin-6-yl]phenyl}urea, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Edwards, T, Abendroth, J, Chun, L. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of RAF Isoforms and Active Dimers by LY3009120 Leads to Anti-tumor Activities in RAS or BRAF Mutant Cancers.
Cancer Cell, 28, 2015
|
|
7DD5
 
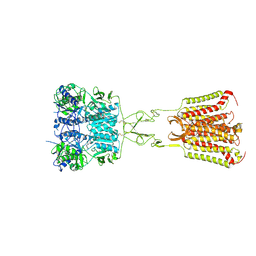 | | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD7
 
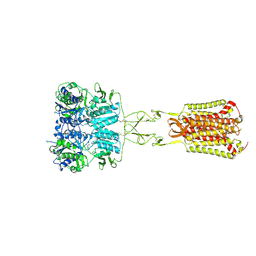 | | Structure of Calcium-Sensing Receptor in complex with Evocalcet | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD6
 
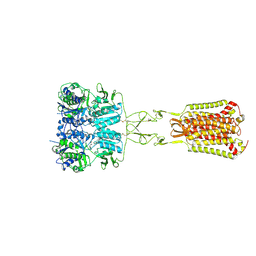 | | Structure of Ca2+/L-Trp-bonnd Calcium-Sensing Receptor in active state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DSC
 
 | | CALHM1 open state with disordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
7DSD
 
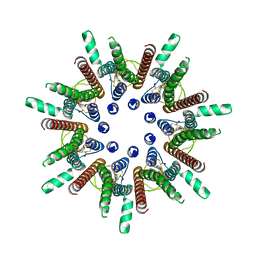 | | CALHM1 close state with disordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
7DSE
 
 | | CALHM1 close state with ordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
8IP6
 
 | | Cryo-EM structure of hMRS2-rest | | Descriptor: | CHLORIDE ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP3
 
 | | Cryo-EM structure of hMRS2-Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
