3TTI
 
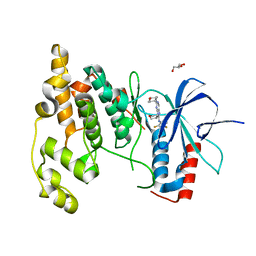 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1D5Q
 
 | | SOLUTION STRUCTURE OF A MINI-PROTEIN REPRODUCING THE CORE OF THE CD4 SURFACE INTERACTING WITH THE HIV-1 ENVELOPE GLYCOPROTEIN | | Descriptor: | CHIMERIC MINI-PROTEIN | | Authors: | Vita, C, Drakopoulou, E, Vizzanova, J, Rochette, S, Martin, L, Menez, A, Roumestand, C, Yang, Y.S, Ylisastigui, L, Benjouad, A, Gluckman, J.C. | | Deposit date: | 1999-10-11 | | Release date: | 2000-10-11 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2SNV
 
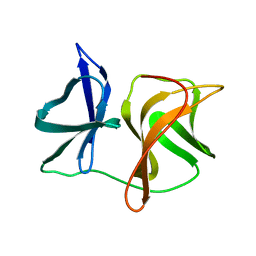 | |
3ISU
 
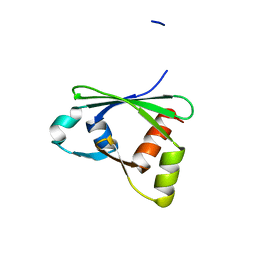 | | Crystal structure of the RGC domain of IQGAP3 | | Descriptor: | Ras GTPase-activating-like protein IQGAP3 | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the RGC domain of IQGAP3
to be published
|
|
6RMD
 
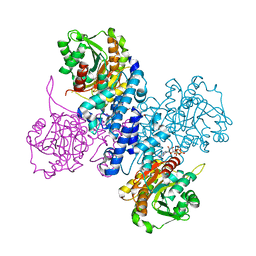 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
3IS2
 
 | | 2.3 Angstrom Crystal Structure of a Cys71 Sulfenic Acid form of Vivid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Vivid PAS protein VVD | | Authors: | Zoltowski, B.D, Lamb, J.S, Pabit, S.A, Li, L, Pollack, L, Crane, B.R. | | Deposit date: | 2009-08-25 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Illuminating solution responses of a LOV domain protein with photocoupled small-angle X-ray scattering.
J.Mol.Biol., 393, 2009
|
|
1WPO
 
 | | HYDROLYTIC ENZYME HUMAN CYTOMEGALOVIRUS PROTEASE | | Descriptor: | HUMAN CYTOMEGALOVIRUS PROTEASE, SULFATE ION | | Authors: | Tong, L. | | Deposit date: | 1996-07-23 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new serine-protease fold revealed by the crystal structure of human cytomegalovirus protease.
Nature, 383, 1996
|
|
5XVW
 
 | |
5XJN
 
 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7KCF
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-24512 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-hydroxyphenyl)-5-methyl-2-phenyl-3-(piperidin-1-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, GLYCEROL, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KDA
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 34 | | Descriptor: | 2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KDB
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 35 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(4-hydroxyphenyl)-2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KCE
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 2 | | Descriptor: | 5-methyl-2,3-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KCC
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AG-270 | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclohex-1-en-1-yl)-6-(4-methoxyphenyl)-2-phenyl-5-[(pyridin-2-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
5XWP
 
 | | Crystal structure of LbuCas13a-crRNA-target RNA ternary complex | | Descriptor: | RNA (30-MER), RNA (59-MER), Uncharacterized protein | | Authors: | Liu, L, Li, X, Li, Z, Wang, Y. | | Deposit date: | 2017-06-30 | | Release date: | 2017-09-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | The Molecular Architecture for RNA-Guided RNA Cleavage by Cas13a.
Cell, 170, 2017
|
|
5XPD
 
 | |
2YOQ
 
 | | Structure of FAM3B PANDER E30 construct | | Descriptor: | GLYCEROL, PROTEIN FAM3B | | Authors: | Johansson, P, Bernstrom, J, Gorman, T, Oster, L, Backstrom, S, Schweikart, F, Xu, B, Xue, Y, Holmberg Schiavone, L. | | Deposit date: | 2012-10-26 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fam3B Pander and Fam3C Ilei Represent a Distinct Class of Signaling Molecules with a Non-Cytokine-Like Fold.
Structure, 21, 2013
|
|
6SCR
 
 | |
7K5V
 
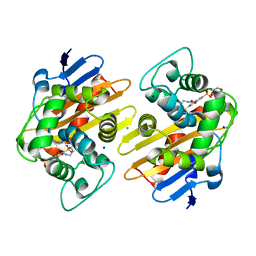 | | OXA-48 bound by Compound 3.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-09-17 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
7JVF
 
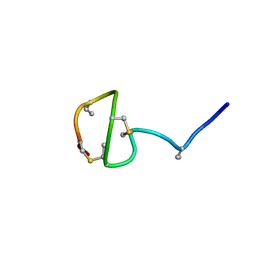 | |
7K7A
 
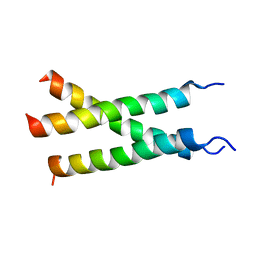 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
3EF7
 
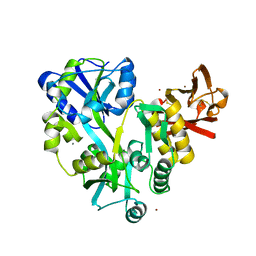 | |
5Y21
 
 | |
3ID8
 
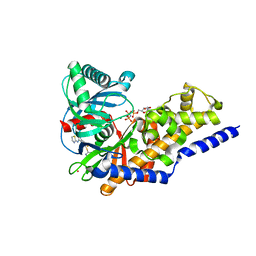 | | Ternary complex of human pancreatic glucokinase crystallized with activator, glucose and AMP-PNP | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, Glucokinase, MAGNESIUM ION, ... | | Authors: | Petit, P, Gluais, L, Lagarde, A, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2009-07-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6Y7N
 
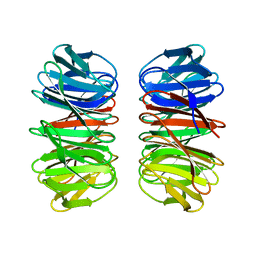 | | The crystal structure of the eight-bladed symmetrical designer protein Tako8 in the presence of tellurotungstic Anderson-Evans (TEW) | | Descriptor: | Tako8 | | Authors: | Vandebroek, L, Noguchi, H, Parac-Vogt, T.N, Van Meervelt, L, Voet, A.R.D. | | Deposit date: | 2020-03-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Shape and Size Complementarity-Induced Formation of Supramolecular Protein Assemblies with Metal-Oxo Clusters
Cryst.Growth Des., 2021
|
|
