8EV2
 
 | | Dual Modulators | | Descriptor: | (3aS,4R,9bR)-4-(2-chloro-4-hydroxyphenyl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[c]quinoline-8-sulfonamide, (3~{a}~{R},4~{S},9~{b}~{S})-4-(2-chloranyl-4-oxidanyl-phenyl)-2,3,3~{a},4,5,9~{b}-hexahydro-1~{H}-cyclopenta[c]quinoline-8-sulfonamide, Estrogen receptor, ... | | Authors: | Tinivella, A, Nwachukwu, J.C, Angeli, A, Foschi, F, Benatti, A.L, Pinzi, L, Izard, T, Ferraroni, M, Rangarajan, E.S, Christodoulou, M, Passarella, D, Supuran, C, Nettles, K.W, Rastelli, G. | | Deposit date: | 2022-10-19 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design, synthesis, biological evaluation and crystal structure determination of dual modulators of carbonic anhydrases and estrogen receptors.
Eur.J.Med.Chem., 246, 2022
|
|
6HUE
 
 | | ParkinS65N | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | McWilliams, T.G, Barini, E, Pohjolan-Pirhonen, R, Brooks, S.P, Singh, F, Burel, S, Balk, K, Kumar, A, Montava-Garriga, L, Prescott, A.R, Hassoun, S.M, Mouton-Liger, F, Ball, G, Hills, R, Knebel, A, Ulusoy, A, Di Monte, D.A, Tamjar, J, Antico, O, Fears, K, Smith, L, Brambilla, R, Palin, E, Valori, M, Eerola-Rautio, J, Tienari, P, Corti, O, Dunnett, S.B, Ganley, I.G, Suomalainen, A, Muqit, M.M.K. | | Deposit date: | 2018-10-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Phosphorylation of Parkin at serine 65 is essential for its activation in vivo .
Open Biology, 8, 2018
|
|
5Z32
 
 | | LPS bound solution NMR structure of WS2-VR18 | | Descriptor: | VAL-ALA-ARG-GLY-TRP-GLY-ARG-LYS-CYS-PRO-LEU-PHE-GLY-LYS-ASN-LYS-SER-ARG | | Authors: | Bhunia, A, Mohid, S.A. | | Deposit date: | 2018-01-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Application of tungsten disulfide quantum dot-conjugated antimicrobial peptides in bio-imaging and antimicrobial therapy.
Colloids Surf B Biointerfaces, 176, 2019
|
|
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
5DK6
 
 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
7CU2
 
 | | CRYSTAL STRUCTURE OF STREPTOMYCES ALBOGRISEOLUS FLAVIN-DEPENDENT TRYPTOPHAN 6-HALOGENASE THAL IN COMPLEX WITH REDUCED FAD | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
7CU0
 
 | | Crystal structure of Streptomyces albogriseolus flavin-dependent tryptophan 6-halogenase Thal in complex with tryptophan | | Descriptor: | TRYPTOPHAN, Tryptophan 6-halogenase | | Authors: | Chitnumsub, P, Jaruwat, A, Phintha, A, Chaiyen, P. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dissecting the low catalytic capability of flavin-dependent halogenases.
J.Biol.Chem., 296, 2020
|
|
8EXA
 
 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
5XWU
 
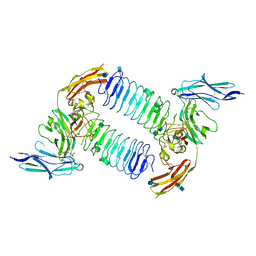 | | Crystal structure of PTPdelta Ig1-Ig3 in complex with SALM2 LRR-Ig | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.162 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
7VQH
 
 | |
8EX9
 
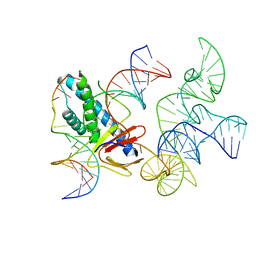 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 2 (RuvC domain unresolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
7S4B
 
 | |
7S3S
 
 | |
7S3K
 
 | |
7M78
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7M75
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
7VRJ
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF Allochromatium tepidum | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Tani, K, Kobayashi, K, Hosogi, N, Ji, X.-C, Nagashima, S, Nagashima, K.V.P, Tsukatani, Y, Kanno, R, Hall, M, Yu, L.-J, Ishikawa, I, Okura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-10-23 | | Release date: | 2022-05-04 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | A Ca 2+ -binding motif underlies the unusual properties of certain photosynthetic bacterial core light-harvesting complexes.
J.Biol.Chem., 298, 2022
|
|
7M76
 
 | | Room Temperature XFEL Crystallography reveals asymmetry in the vicinity of the two phylloquinones in Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Keable, S.M, Simon, P.S, Kolsch, A, Kern, J, Yachandra, V.K, Zouni, A, Yano, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I.
Sci Rep, 11, 2021
|
|
8TBP
 
 | | HLA-DRB1*15:01 in complex with smith antigen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ting, Y.T, Broury, A, Ooi, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.12621117 Å) | | Cite: | Smith-specific regulatory T cells halt the progression of lupus nephritis.
Nat Commun, 15, 2024
|
|
6I8T
 
 | |
7A8U
 
 | | Crystal structure of sarcomeric protein FATZ-1 (d91-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.802 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
7A8T
 
 | | Crystal structure of sarcomeric protein FATZ-1 (mini-FATZ-1 construct) in complex with rod domain of alpha-actinin-2 | | Descriptor: | Alpha-actinin-2, Myozenin-1 | | Authors: | Sponga, A, Arolas, J.L, Rodriguez Chamorro, A, Mlynek, G, Hollerl, E, Schreiner, C, Pedron, M, Kostan, J, Ribeiro, E.A, Djinovic-Carugo, K. | | Deposit date: | 2020-08-31 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Order from disorder in the sarcomere: FATZ forms a fuzzy but tight complex and phase-separated condensates with alpha-actinin.
Sci Adv, 7, 2021
|
|
4ZZZ
 
 | | Structure of human PARP1 catalytic domain bound to an isoindolinone inhibitor | | Descriptor: | 2-(3-methoxypropyl)-3-oxo-2,3-dihydro-1H-isoindole-4-carboxamide, GLYCEROL, POLY [ADP-RIBOSE] POLYMERASE 1, ... | | Authors: | Casale, E, Fasolini, M, Papeo, G, Posteri, H, Borghi, D, Busel, A.A, Caprera, F, Ciomei, M, Cirla, A, Corti, E, DAnello, M, Fasolini, M, Felder, E.R, Forte, B, Galvani, A, Isacchi, A, Khvat, A, Krasavin, M.Y, Lupi, R, Orsini, P, Perego, R, Pesenti, E, Pezzetta, D, Rainoldi, S, RiccardiSirtori, F, Scolaro, A, Sola, F, Zuccotto, F, Donati, D, Montagnoli, A. | | Deposit date: | 2015-04-15 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 2-[1-(4,4-Difluorocyclohexyl)Piperidin-4-Yl]-6-Fluoro-3-Oxo-2,3-Dihydro-1H-Isoindole-4-Carboxamide (Nms-P118): A Potent, Orally Available and Highly Selective Parp- 1 Inhibitor for Cancer Therapy.
J.Med.Chem., 58, 2015
|
|
5XWT
 
 | | Crystal structure of PTPdelta Ig1-Fn1 in complex with SALM5 LRR-Ig | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goto-Ito, S, Yamagata, A, Sato, Y, Fukai, S. | | Deposit date: | 2017-06-30 | | Release date: | 2018-06-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4.178 Å) | | Cite: | Structural basis of trans-synaptic interactions between PTP delta and SALMs for inducing synapse formation.
Nat Commun, 9, 2018
|
|
5LOF
 
 | | Crystal structure of the MBP-MCL1 complex with highly selective and potent inhibitor of MCL1 | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(5-fluoranylfuran-2-yl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-[2,2,2-tris(fluoranyl)ethyl]pyrazol-3-yl]methoxy]phenyl]propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Kotschy, A, Szlavik, Z, Murray, J, Davidson, J, Csekei, M, Paczal, A, Szabo, Z, Sipos, S, Radics, G, Proszenyak, A, Balint, B, Ondi, L, Blasko, G, Robertson, A, Surgenor, A, Chen, I, Matassova, N, Smith, J, Pedder, C, Graham, C, Geneste, O. | | Deposit date: | 2016-08-09 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models.
Nature, 538, 2016
|
|
