8G3E
 
 | | Crystal structure of human WDR5 in complex with (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide (compound 2, WDR5-MYC inhibitor) | | Descriptor: | (1M)-N-[(3,5-difluoro[1,1'-biphenyl]-4-yl)methyl]-6-methyl-4-oxo-1-(pyridin-3-yl)-1,4-dihydropyridazine-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Zhao, M. | | Deposit date: | 2023-02-07 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery and Structure-Based Design of Inhibitors of the WD Repeat-Containing Protein 5 (WDR5)-MYC Interaction.
J.Med.Chem., 66, 2023
|
|
4ZHJ
 
 | |
6DE7
 
 | |
8F1G
 
 | | Crystal structure of human WDR5 in complex with compound WM662 | | Descriptor: | (2S)-2-({(2S)-3-(3'-chloro[1,1'-biphenyl]-4-yl)-1-oxo-1-[(1H-tetrazol-5-yl)amino]propan-2-yl}oxy)propanoic acid, GLYCEROL, SULFATE ION, ... | | Authors: | Liu, H. | | Deposit date: | 2022-11-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of Potent Small-Molecule Inhibitors of WDR5-MYC Interaction.
Acs Chem.Biol., 18, 2023
|
|
8IA3
 
 | | Crystal structure of human USF2 bHLHLZ domain in complex with DNA | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*GP*TP*CP*AP*CP*GP*TP*GP*CP*CP*CP*GP*TP*C)-3'), DNA (5'-D(P*GP*AP*CP*GP*GP*GP*CP*AP*CP*GP*TP*GP*AP*CP*GP*CP*GP*C)-3'), Upstream stimulatory factor 2 | | Authors: | Huang, C, Fang, P, Wang, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Tetramerization of upstream stimulating factor USF2 requires the elongated bent leucine zipper of the bHLH-LZ domain.
J.Biol.Chem., 299, 2023
|
|
6EB0
 
 | | STRUCTURE OF 4-HYDROXYPHENYLACETATE 3-MONOOXYGENASE (HPAB), OXYGENASE COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | 4-hydroxyphenylacetate 3-monooxygenase, oxygenase subunit, ACETATE ION | | Authors: | Zhou, D, Kandavelu, P, Zhang, H, Wang, B.C, Yan, Y, Rose, J. | | Deposit date: | 2018-08-03 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Insights into Catalytic Versatility of the Flavin-dependent Hydroxylase (HpaB) from Escherichia coli.
Sci Rep, 9, 2019
|
|
6LHC
 
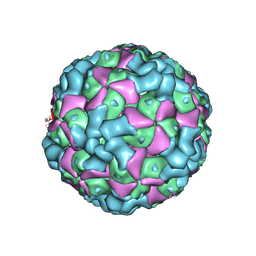 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
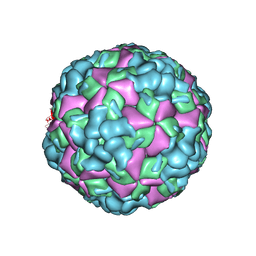 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
5FFC
 
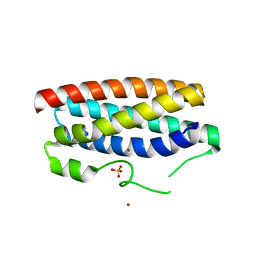 | | CopM in the Cu(II)-bound form | | Descriptor: | COPPER (II) ION, CopM, SULFATE ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFE
 
 | | CopM in the Ag-bound form (by soaking) | | Descriptor: | CopM, SILVER ION | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFD
 
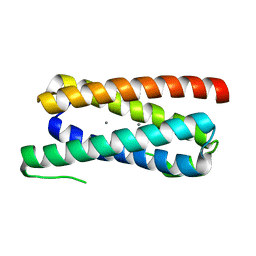 | |
5FEJ
 
 | | CopM in the Cu(I)-bound form | | Descriptor: | COPPER (I) ION, CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-17 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5FFA
 
 | |
5FFB
 
 | | CopM in the apo form | | Descriptor: | CopM | | Authors: | Zhao, S, Wang, X, Liu, L. | | Deposit date: | 2015-12-18 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural basis for copper/silver binding by the Synechocystis metallochaperone CopM.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2IBX
 
 | | Influenza virus (VN1194) H5 HA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yamada, S, Russell, R.J, Gamblin, S.J, Skehel, J.J, Kawaoka, Y. | | Deposit date: | 2006-09-12 | | Release date: | 2006-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors.
Nature, 444, 2006
|
|
4LOC
 
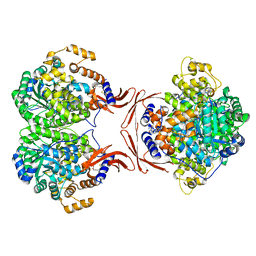 | |
4M6V
 
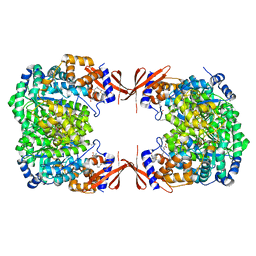 | |
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
5WYS
 
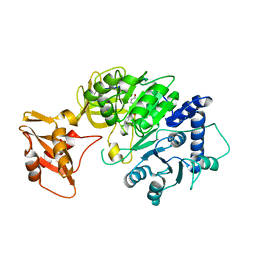 | | luciferase with inhibitor 3i | | Descriptor: | 5-[(3R)-3-(4-boranylphenyl)-3-oxidanyl-propyl]-2-oxidanyl-benzoic acid, Luciferin 4-monooxygenase | | Authors: | Gu, L, Su, J, Wang, F. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Inhibiting Firefly Bioluminescence by Chalcones
Anal. Chem., 89, 2017
|
|
6T6P
 
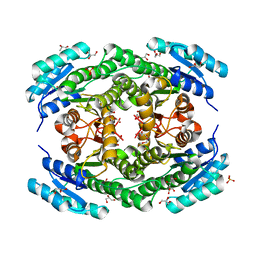 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) at 1.57 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
8RUC
 
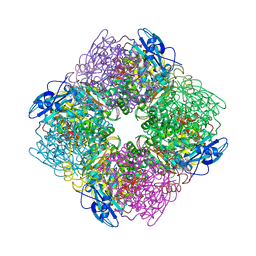 | | ACTIVATED SPINACH RUBISCO COMPLEXED WITH 2-CARBOXYARABINITOL BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RIBULOSE-1,5-BISPHOSPHATE CARBOXYLASE/OXYGENASE | | Authors: | Andersson, I, Knight, S, Branden, C.-I. | | Deposit date: | 1996-02-22 | | Release date: | 1996-08-01 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large structures at high resolution: the 1.6 A crystal structure of spinach ribulose-1,5-bisphosphate carboxylase/oxygenase complexed with 2-carboxyarabinitol bisphosphate.
J.Mol.Biol., 259, 1996
|
|
6T6N
 
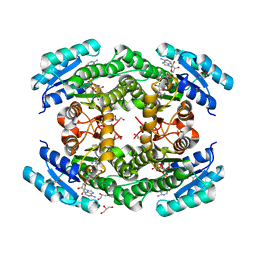 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) in complex with NADH at 2.5 A resolution | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-[acyl-carrier protein] reductase, D-MALATE, ... | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
