4P5Q
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(2-chlorophenyl)-N-(3-ethoxypropyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-19 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4P5Z
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-[4-({[3-(trifluoromethyl)phenyl]carbamoyl}amino)phenyl]-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-20 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4P4C
 
 | | Human EphA3 Kinase domain in complex with quinoxaline derivatives | | Descriptor: | 2-amino-1-(3-methoxyphenyl)-1H-pyrrolo[2,3-b]quinoxaline-3-carboxamide, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Pyrrolo[3,2-b]quinoxaline Derivatives as Types I1/2 and II Eph Tyrosine Kinase Inhibitors: Structure-Based Design, Synthesis, and in Vivo Validation.
J.Med.Chem., 57, 2014
|
|
4TWO
 
 | | Human EphA3 Kinase domain in complex with compound 164 | | Descriptor: | 5-{[3-carbamoyl-4-(3,4-dimethylphenyl)-5-methylthiophen-2-yl]amino}-5-oxopentanoic acid, Ephrin type-A receptor 3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2014-07-01 | | Release date: | 2015-05-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural Analysis of the Binding of Type I, I1/2, and II Inhibitors to Eph Tyrosine Kinases.
Acs Med.Chem.Lett., 6, 2015
|
|
7QHD
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QHE
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
3EUO
 
 | |
3EUQ
 
 | |
3EUT
 
 | |
3CHT
 
 | |
3CHU
 
 | | Crystal Structure of Di-iron Aurf | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3F5O
 
 | | Crystal Structure of hTHEM2(undecan-2-one-CoA) complex | | Descriptor: | CHLORIDE ION, COENZYME A, HEXAETHYLENE GLYCOL, ... | | Authors: | Xu, H, Gong, W. | | Deposit date: | 2008-11-04 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis
Biochemistry, 48, 2009
|
|
4JND
 
 | | Structure of a C.elegans sex determining protein | | Descriptor: | Ca(2+)/calmodulin-dependent protein kinase phosphatase, MAGNESIUM ION | | Authors: | Feng, Y, Zhang, Y, Ge, J, Yang, M. | | Deposit date: | 2013-03-15 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.652 Å) | | Cite: | Structural insight into Caenorhabditis elegans sex-determining protein FEM-2.
J.Biol.Chem., 288, 2013
|
|
4K4A
 
 | | X-ray crystal structure of E. coli YdiI complexed with phenacyl-CoA | | Descriptor: | Esterase YdiI, phenacyl coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4B
 
 | | X-ray crystal structure of E. coli YdiI complexed with undeca-2-one-CoA | | Descriptor: | CHLORIDE ION, Esterase YdiI, undeca-2-one coenzyme A | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
4K4D
 
 | | X-ray crystal structure of E. coli YbdB complexed with 2,4-dihydroxyphenacyl-CoA | | Descriptor: | 2,4-dihydroxyphenacyl coenzyme A, ACETATE ION, MALONATE ION, ... | | Authors: | Ru, W, Farelli, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2013-04-12 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Catalysis in the Escherichia coli Hotdog-fold Thioesterase Paralogs YdiI and YbdB.
Biochemistry, 53, 2014
|
|
8X5D
 
 | |
8WFX
 
 | | Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii | | Descriptor: | CRISPR system Cms endoribonuclease Csm3, CRISPR system Cms protein Csm2, CRISPR system Cms protein Csm4, ... | | Authors: | Huo, Y, Ma, X, Jiang, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Type-III-A structure of mycobacteria CRISPR-Csm complexes involving atypical crRNAs.
Int.J.Biol.Macromol., 260, 2024
|
|
4NQI
 
 | | Structure of the N-terminal I-BAR domain (1-259) of D.Discoideum IBARa | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SH3 domain-containing protein | | Authors: | Witte, G, Faix, J, Runge-Wollmann, P. | | Deposit date: | 2013-11-25 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The inverse BAR domain protein IBARa drives membrane remodeling to control osmoregulation, phagocytosis and cytokinesis.
J.Cell.Sci., 127, 2014
|
|
4MKI
 
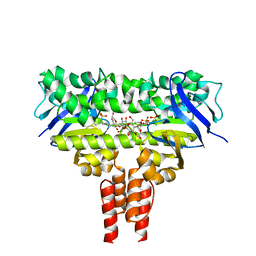 | | Cobalt transporter ATP-binding subunit | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | Authors: | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
3M6I
 
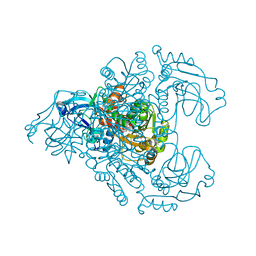 | | L-arabinitol 4-dehydrogenase | | Descriptor: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
7SWW
 
 | |
7SWX
 
 | |
7SIU
 
 | | Crystal structure of HPK1 (MAP4K1) complex with inhibitor A-745 | | Descriptor: | 6-[(5R)-5-benzamidocyclohex-1-en-1-yl]-3-[(1-methyl-1H-pyrazol-4-yl)amino]pyrazine-2-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1, SULFATE ION | | Authors: | Longenecker, K.L, Korepanova, A, Qiu, W. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | The HPK1 Inhibitor A-745 Verifies the Potential of Modulating T Cell Kinase Signaling for Immunotherapy.
Acs Chem.Biol., 17, 2022
|
|
3HWP
 
 | | Crystal structure and computational analyses provide insights into the catalytic mechanism of 2, 4-diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PhlG, ... | | Authors: | He, Y.-X, Huang, L, Xue, Y, Fei, X, Teng, Y.-B, Zhou, C.-Z. | | Deposit date: | 2009-06-18 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Computational Analyses Provide Insights into the Catalytic Mechanism of 2,4-Diacetylphloroglucinol Hydrolase PhlG from Pseudomonas fluorescens.
J.Biol.Chem., 285, 2010
|
|
