3ZNS
 
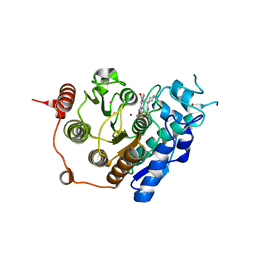 | | HDAC7 bound with TFMO inhibitor tmp942 | | Descriptor: | HISTONE DEACETYLASE 7, N-{[1-methyl-4-(4-phenyl-1,3-thiazol-2-yl)piperidin-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | Authors: | Lobera, M, Madauss, K.P, Pohlhaus, D.T, Trump, R.P, Nolan, M.A. | | Deposit date: | 2013-02-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
3ZNR
 
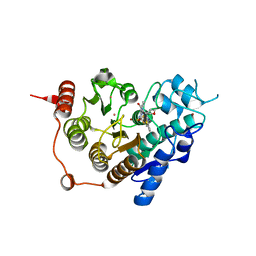 | | HDAC7 bound with inhibitor TMP269 | | Descriptor: | HISTONE DEACETYLASE 7, N-{[4-(4-phenyl-1,3-thiazol-2-yl)tetrahydro-2H-pyran-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | Authors: | Lobera, m, madauss, k, pohlhaus, d, trump, r, nolan, m. | | Deposit date: | 2013-02-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
6IE1
 
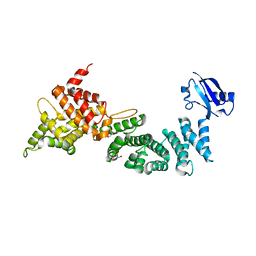 | | Crystal Structure of ELMO2(Engulfment and cell motility protein 2) | | Descriptor: | Engulfment and cell motility protein 2, GLYCEROL | | Authors: | Weng, Z.F, Lin, L, Zhang, R.G, Zhu, J.W. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of BAI1/ELMO2 complex reveals an action mechanism of adhesion GPCRs via ELMO family scaffolds
Nat Commun, 10, 2019
|
|
8HMV
 
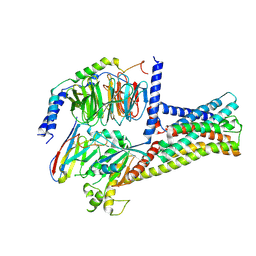 | | Structure of GPR21-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Wong, T.S, Gao, W. | | Deposit date: | 2022-12-05 | | Release date: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of orphan G protein-coupled receptor GPR21.
MedComm (2020), 4, 2023
|
|
5UU7
 
 | | Tetragonal thermolysin (295 K) in the presence of 50% mpd | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60001385 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UUE
 
 | |
5UU9
 
 | |
6IDX
 
 | | Crystal Structure of BAI1/ELMO2 complex | | Descriptor: | Adhesion G protein-coupled receptor B1, Engulfment and cell motility protein 2 | | Authors: | Weng, Z.F, Lin, L, Zhu, J.W, Zhang, R.G. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structure of BAI1/ELMO2 complex reveals an action mechanism of adhesion GPCRs via ELMO family scaffolds
Nat Commun, 10, 2019
|
|
7DVA
 
 | | Structure of wild type Bt4394, a GH20 family sulfoglycosidase, in complex with 6S-GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, GLYCEROL | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DUP
 
 | | Apo structure of wild type Bt4394, a GH20 family sulfoglycosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-10 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
7DVB
 
 | | D335N variant of Bt4394 in complex with 6SO3-NAG-oxazoline intermediate | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, [(3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-methyl-6,7-bis(oxidanyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]oxazol-1-ium-5-yl]methyl sulfate | | Authors: | Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-01-13 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanistic and Structural Insights into the Specificity and Biological Functions of Bacterial Sulfoglycosidases
Acs Catalysis, 13, 2023
|
|
8IJA
 
 | | Cryo-EM structure of human HCAR2-Gi complex with niacin | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-26 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJD
 
 | | Cryo-EM structure of human HCAR2-Gi complex with MK-6892 | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJ3
 
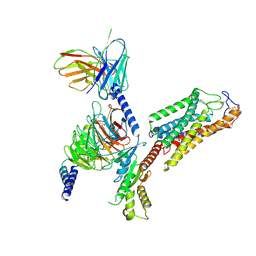 | | Cryo-EM structure of human HCAR2-Gi complex without ligand (apo state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
8IJB
 
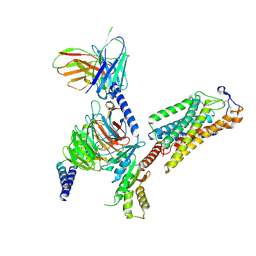 | | Cryo-EM structure of human HCAR2-Gi complex with acipimox | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Pan, X, Fang, Y. | | Deposit date: | 2023-02-27 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural insights into ligand recognition and selectivity of the human hydroxycarboxylic acid receptor HCAR2.
Cell Discov, 9, 2023
|
|
5UUC
 
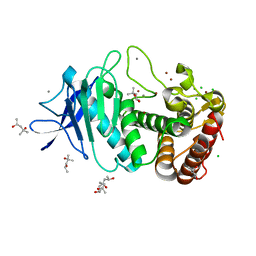 | | Tetragonal thermolysin cryocooled to 100 K with 50% mpd as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60000932 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UU8
 
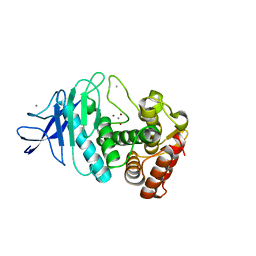 | |
5UUD
 
 | | Tetragonal thermolysin cryocooled to 100 K with 50% dmf as cryoprotectant | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYLFORMAMIDE, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60000348 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5UY8
 
 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | 5-[(5S)-5-ethyl-5-methyl-6-oxo-1,4,5,6-tetrahydropyridin-3-yl]-N-(6-fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)thiophene-2-sulfonamide, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, ... | | Authors: | Wang, J, Wang, Y, Fales, K.R, Atwell, S, Clawson, D. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
5UZ0
 
 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, MAGNESIUM ION, ... | | Authors: | Atwell, S, Wang, Y, Fales, K.R, Clawson, D, Wang, J. | | Deposit date: | 2017-02-24 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
5UUA
 
 | |
8IOY
 
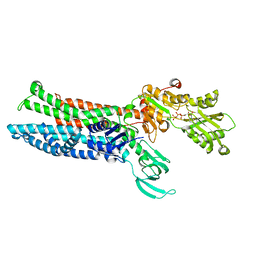 | | Structure of ATP7B C983S/C985S/D1027A mutant with AMP-PNP | | Descriptor: | Copper-transporting ATPase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yang, G, Xu, L, Guo, J, Wu, Z. | | Deposit date: | 2023-03-13 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
5UUB
 
 | |
5UN3
 
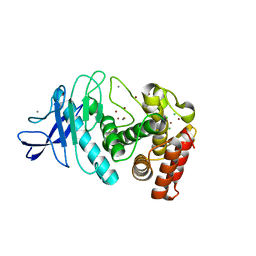 | | Tetragonal thermolysin (295 K) in the presence of 50% xylose | | Descriptor: | CALCIUM ION, CHLORIDE ION, Thermolysin, ... | | Authors: | Juers, D. | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.60004437 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7D59
 
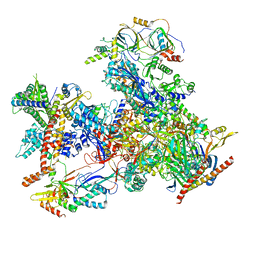 | | cryo-EM structure of human RNA polymerase III in apo state | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Wang, Q, Wan, F, Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into transcriptional regulation of human RNA polymerase III.
Nat.Struct.Mol.Biol., 28, 2021
|
|
