5BPE
 
 | | Crystal structure of EV71 3Cpro in complex with a potent and selective Inhibitor | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{S})-1-cyano-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-3-phenyl-2-[[(~{E})-3-phenylprop-2-enoyl]amino]propanamide, EV71 3Cpro | | Authors: | Luqing, S, Yin, Z. | | Deposit date: | 2015-05-28 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanohydrin as an Anchoring Group for Potent and Selective Inhibitors of Enterovirus 71 3C Protease
J.Med.Chem., 58, 2015
|
|
5H25
 
 | | EED in complex with PRC2 allosteric inhibitor compound 11 | | Descriptor: | 5-(2-fluorophenyl)-2,3-dihydroimidazo[2,1-a]isoquinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
1M5B
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH 2-Me-Tet-AMPA AT 1.85 A RESOLUTION. | | Descriptor: | (S)-2-AMINO-3-[3-HYDROXY-5-(2-METHYL-2H-TETRAZOL-5-YL)ISOXAZOL-4-YL]PROPIONIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
3JBH
 
 | | TWO HEAVY MEROMYOSIN INTERACTING-HEADS MOTIFS FLEXIBLE DOCKED INTO TARANTULA THICK FILAMENT 3D-MAP ALLOWS IN DEPTH STUDY OF INTRA- AND INTERMOLECULAR INTERACTIONS | | Descriptor: | MYOSIN 2 ESSENTIAL LIGHT CHAIN STRIATED MUSCLE, MYOSIN 2 HEAVY CHAIN STRIATED MUSCLE, MYOSIN 2 REGULATORY LIGHT CHAIN STRIATED MUSCLE | | Authors: | Alamo, L, Qi, D, Wriggers, W, Pinto, A, Zhu, J, Bilbao, A, Gillilan, R.E, Hu, S, Padron, R. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis.
J. Mol. Biol., 428, 2016
|
|
1M5E
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH ACPA AT 1.46 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1MS7
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.97 A resolution, Crystallization in the presence of zinc acetate | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, Glutamate receptor subunit 2, ZINC ION | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-19 | | Release date: | 2003-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | GluR2 ligand-binding core complexes: importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists
FEBS Lett., 531, 2002
|
|
1N0T
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | Descriptor: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | Deposit date: | 2002-10-15 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
8T5C
 
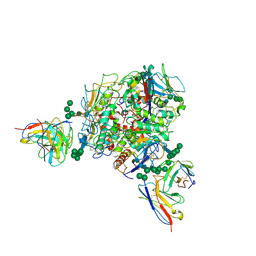 | | Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8.11G Heavy Chain, 8.11G Light Chain, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Cleavage-intermediate Lassa virus trimer elicits neutralizing responses, identifies neutralizing nanobodies, and reveals an apex-situated site-of-vulnerability.
Nat Commun, 15, 2024
|
|
1NNP
 
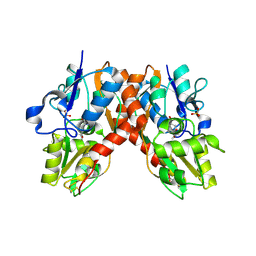 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | Authors: | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1NNK
 
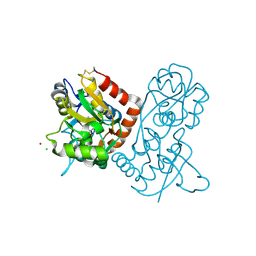 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Lunn, M.-L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
1M5C
 
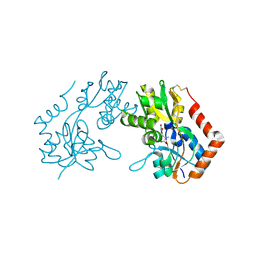 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH Br-HIBO AT 1.65 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1MQD
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-Des-Me-AMPA at 1.46 A resolution. Crystallization in the presence of lithium sulfate. | | Descriptor: | (S)-2-AMINO-3-(3-HYDROXY-ISOXAZOL-4-YL)PROPIONIC ACID, GLYCEROL, Glutamate receptor subunit 2, ... | | Authors: | Kasper, C, Lunn, M.-L, Liljefors, T, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2002-09-16 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | GluR2 ligand-binding core complexes: Importance of the isoxazolol moiety and 5-substituent for the binding mode of AMPA-type agonists.
FEBS Lett., 531, 2002
|
|
1M5D
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH Br-HIBO AT 1.73 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(4-BROMO-3-HYDROXY-ISOXAZOL-5-YL)PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
1M5F
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J-Y702F) IN COMPLEX WITH ACPA AT 1.95 A RESOLUTION | | Descriptor: | (S)-2-AMINO-3-(3-CARBOXY-5-METHYLISOXAZOL-4-YL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | Authors: | Hogner, A, Kastrup, J.S, Jin, R, Liljefors, T, Mayer, M.L, Egebjerg, J, Larsen, I.K, Gouaux, E. | | Deposit date: | 2002-07-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Basis for AMPA Receptor Activation and Ligand Selectivity:
Crystal Structures of Five Agonist Complexes with the GluR2 Ligand-binding
Core
J.Mol.Biol., 322, 2002
|
|
2I3V
 
 | | Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor: Structure of G725C mutant | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, ZINC ION | | Authors: | Armstrong, N, Jasti, J, Beich-Frandsen, M, Gouaux, E. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Measurement of Conformational Changes accompanying Desensitization in an Ionotropic Glutamate Receptor.
Cell(Cambridge,Mass.), 127, 2006
|
|
5H24
 
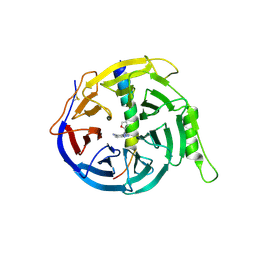 | | EED in complex with PRC2 allosteric inhibitor compound 8 | | Descriptor: | 5-(furan-2-ylmethylamino)-[1,2,4]triazolo[4,3-a]pyridine-6-carbonitrile, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of First-in-Class, Potent, and Orally Bioavailable Embryonic Ectoderm Development (EED) Inhibitor with Robust Anticancer Efficacy
J. Med. Chem., 60, 2017
|
|
5T92
 
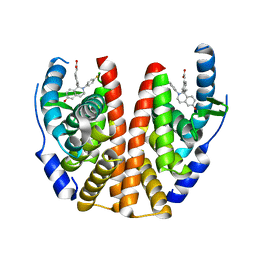 | | ESTROGEN RECEPTOR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH (2E)-3-{4-[(1R)-2-(4-fluorophenyl)-6-hydroxy-1-methy l-1,2,3,4- tetrahydroisoquinolin-1-yl]phenyl}prop-2-enoic acid | | Descriptor: | (2E)-3-{4-[(1R)-2-(4-fluorophenyl)-6-hydroxy-1-methyl-1,2,3,4-tetrahydroisoquinolin-1-yl]phenyl}prop-2-enoic acid, Estrogen receptor | | Authors: | Kirby, C, Baird, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Discovery of an Acrylic Acid Based Tetrahydroisoquinoline as an Orally Bioavailable Selective Estrogen Receptor Degrader for ER alpha + Breast Cancer.
J. Med. Chem., 60, 2017
|
|
5T97
 
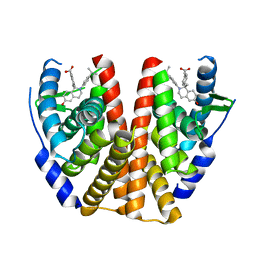 | | ESTROGEN RECEPTOR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH (2E)-3-(4-{(1R)-6-hydroxy-1-methyl-2-[4-(propan-2 -yl)phenyl]-1,2,3,4- tetrahydroisoquinolin-1-yl}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{(1R)-6-hydroxy-1-methyl-2-[4-(propan-2-yl)phenyl]-1,2,3,4-tetrahydroisoquinolin-1-yl}phenyl)prop-2-enoic acid, Estrogen receptor | | Authors: | Kirby, C.A, Baird, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of an Acrylic Acid Based Tetrahydroisoquinoline as an Orally Bioavailable Selective Estrogen Receptor Degrader for ER alpha + Breast Cancer.
J. Med. Chem., 60, 2017
|
|
5TIJ
 
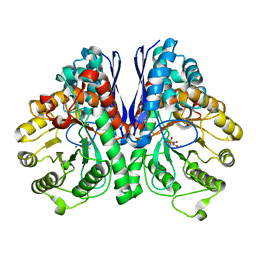 | | Structure of Human Enolase 2 with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonate (purified enantiomer) | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Eradication of ENO1-deleted Glioblastoma through Collateral Lethality
Biorxiv, 2019
|
|
6OZB
 
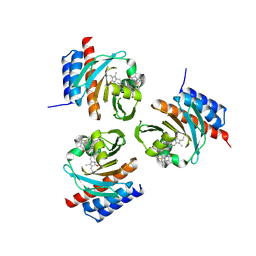 | | Crystal structure of the phycoerythrobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOERYTHROBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5E06
 
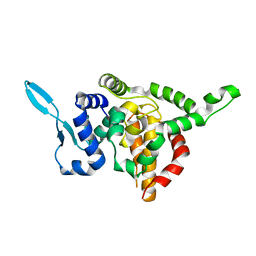 | |
6OZA
 
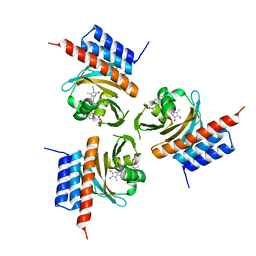 | | Crystal structure of the phycocyanobilin-bound GAF domain from a cyanobacterial phytochrome | | Descriptor: | PHYCOCYANOBILIN, Two-component sensor histidine kinase | | Authors: | Heewhan, S, Xiaoli, Z, Yafang, S, Zhong, R, Wolfgang, G, Kai, H.Z, Xiaojing, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The interplay between chromophore and protein determines the extended excited state dynamics in a single-domain phytochrome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5E05
 
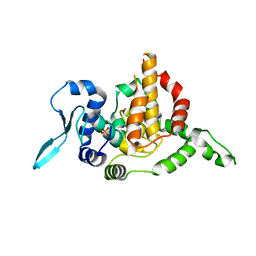 | |
4RLW
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with Butein | | Descriptor: | (2E)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)prop-2-en-1-one, (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
4RLU
 
 | | Crystal Structure of (3R)-hydroxyacyl-ACP dehydratase HadAB hetero-dimer from Mycobacterium tuberculosis complexed with 2',4,4'-trihydroxychalcone | | Descriptor: | (3R)-hydroxyacyl-ACP dehydratase subunit HadA, (3R)-hydroxyacyl-ACP dehydratase subunit HadB, 2',4,4'-TRIHYDROXYCHALCONE, ... | | Authors: | Li, J, Dong, Y, Rao, Z.H. | | Deposit date: | 2014-10-18 | | Release date: | 2015-10-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Molecular basis for the inhibition of beta-hydroxyacyl-ACP dehydratase HadAB complex from Mycobacterium tuberculosis by flavonoid inhibitors.
Protein Cell, 6, 2015
|
|
