7VIW
 
 | | Dark adapted MmCPDII during oxidized to semiquinone TR-SFX studies | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ3
 
 | | class II photolyase MmCPDII oxidized to semiquinone TR-SFX studies (400 us time-point) | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.-C, Nango, E, Ngura Putu, E.P.G, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Hosokawa, Y, Saft, M, Emmerich, H.-J, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Royant, A, Gad, W, Pang, A.H, Chang, C.-W, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
2ZTS
 
 | |
2BDR
 
 | | Crystal Structure of the Putative Ureidoglycolate hydrolase PP4288 from Pseudomonas putida, Northeast Structural Genomics Target PpR49 | | Descriptor: | SODIUM ION, Ureidoglycolate hydrolase | | Authors: | Forouhar, F, Abashidze, M, Jayaraman, S, Ho, C.K, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Putative Ureidoglycolate hydrolase PP4288 from Pseudomonas putida, Northeast Structural Genomics Target PpR49
To be Published
|
|
2BDE
 
 | | Crystal Structure of the cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) from Legionella pneumophila, Northeast Structural Genomics Target LgR1 | | Descriptor: | SULFATE ION, cytosolic IMP-GMP specific 5'-nucleotidase | | Authors: | Forouhar, F, Abashidze, M, Ho, C.K, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) from Legionella pneumophila, Northeast Structural Genomics Target LgR1
To be Published
|
|
2ZUA
 
 | | Crystal structure of nucleoside diphosphate kinase from Haloarcula quadrata | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Ichimura, T, Yamamura, A, Ohtsuka, J, Miyazono, K, Okai, M, Nagata, K, Tanokura, M. | | Deposit date: | 2008-10-15 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Molecular mechanism of distinct salt-dependent enzyme activity of two halophilic nucleoside diphosphate kinases
Biophys.J., 96, 2009
|
|
3A9G
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase apo-form | | Descriptor: | CALCIUM ION, Putative uncharacterized protein, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
2EQJ
 
 | | Solution structure of the TUDOR domain of Metal-response element-binding transcription factor 2 | | Descriptor: | Metal-response element-binding transcription factor 2 | | Authors: | Dang, W, Muto, Y, Isono, K, Watanabe, S, Tarada, T, Kigawa, T, Koseki, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TUDOR domain of Metal-response element-binding transcription factor 2
To be Published
|
|
3A9H
 
 | | Crystal Structure of PQQ-dependent sugar dehydrogenase holo-form | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, Putative uncharacterized protein, ... | | Authors: | Sakuraba, H, Yokono, K, Yoneda, K, Ohshima, T. | | Deposit date: | 2009-10-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic properties and crystal structure of quinoprotein aldose sugar dehydrogenase from hyperthermophilic archaeon Pyrobaculum aerophilum
Arch.Biochem.Biophys., 502, 2010
|
|
3A2E
 
 | | Crystal structure of ginkbilobin-2, the novel antifungal protein from Ginkgo biloba seeds | | Descriptor: | Ginkbilobin-2 | | Authors: | Miyakawa, T, Miyazono, K, Sawano, Y, Hatano, K, Tanokura, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases
Proteins, 77, 2009
|
|
3BIJ
 
 | | Crystal structure of protein GSU0716 from Geobacter sulfurreducens. Northeast Structural Genomics target GsR13 | | Descriptor: | Uncharacterized protein GSU0716 | | Authors: | Forouhar, F, Neely, H, Su, M, Seetharaman, J, Benach, J, Conover, K, Fang, Y, Xiao, R, Owen, L.A, Maglaqui, M, Cunningham, K, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein GSU0716 from Geobacter sulfurreducens.
To be Published
|
|
2F9F
 
 | | Crystal Structure of the Putative Mannosyl Transferase (wbaZ-1)from Archaeoglobus fulgidus, Northeast Structural Genomics Target GR29A. | | Descriptor: | first mannosyl transferase (wbaZ-1) | | Authors: | Zhou, W, Forouhar, F, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-05 | | Release date: | 2006-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Putative Mannosyl Transferase
(wbaZ-1)from Archaeoglobus fulgidus
To be Published
|
|
1J2E
 
 | | Crystal structure of Human Dipeptidyl peptidase IV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV | | Authors: | Hiramatsu, H, Kyono, K, Higashiyama, Y, Fukushima, C, Shima, H, Sugiyama, S, Inaka, K, Yamamoto, A, Shimizu, R. | | Deposit date: | 2002-12-30 | | Release date: | 2003-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure and function of human dipeptidyl peptidase IV, possessing a unique eight-bladed beta-propeller fold.
Biochem.Biophys.Res.Commun., 302, 2003
|
|
2FFI
 
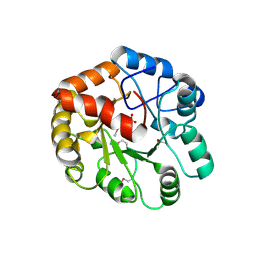 | | Crystal Structure of Putative 2-Pyrone-4,6-Dicarboxylic Acid Hydrolase from Pseudomonas putida, Northeast Structural Genomics Target PpR23. | | Descriptor: | 2-pyrone-4,6-dicarboxylic acid hydrolase, putative, PHOSPHATE ION | | Authors: | Forouhar, F, Su, M, Jayaraman, S, Conover, K, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-12-19 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Putative 2-Pyrone-4,6-Dicarboxylic Acid Hydrolase from Pseudomonas putida, Northeast Structural Genomics Target PpR23.
To be Published
|
|
3OV2
 
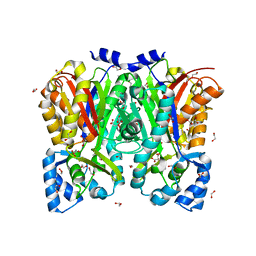 | | Curcumin synthase 1 from Curcuma longa | | Descriptor: | 1,2-ETHANEDIOL, Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
3OV3
 
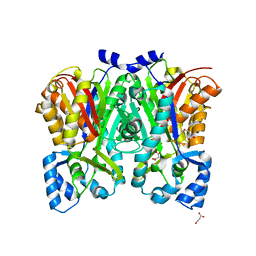 | | G211F mutant of curcumin synthase 1 from Curcuma longa | | Descriptor: | Curcumin synthase, MALONATE ION | | Authors: | Katsuyama, Y, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A hydrophobic cavity discovered in a curcumin synthase facilitates utilization of a beta-keto acid as an extender substrate for the atypical type III polyleteide synthase
To be Published
|
|
7CFA
 
 | |
7CO1
 
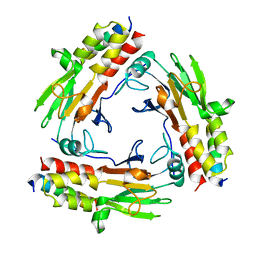 | | Crystal structure of SMAD2 in complex with wild-type CBP | | Descriptor: | CREB-binding protein, Mothers against decapentaplegic homolog 2 | | Authors: | Miyazono, K, Wada, H, Ito, T, Tanokura, M. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-beta signaling.
Sci.Signal., 13, 2020
|
|
8XCD
 
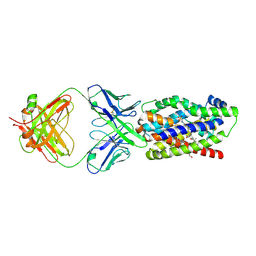 | | Macaca fascicularis NTCP in complex with YN69083 Fab | | Descriptor: | Solute carrier family 10 member a1, TAUROCHOLIC ACID, YN69083 Fab Heavy chain, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-12-08 | | Release date: | 2024-11-13 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for hepatitis B virus restriction by a viral receptor homologue.
Nat Commun, 15, 2024
|
|
7WAB
 
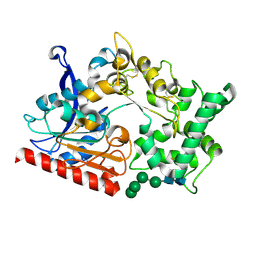 | | Crystal structure of the prolyl endoprotease, PEP, from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPASS (Complex proteins associated with Set1p) component shg1 family protein, ... | | Authors: | Miyazono, K, Kubota, K, Takahashi, K, Tanokura, M. | | Deposit date: | 2021-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure and substrate recognition mechanism of the prolyl endoprotease PEP from Aspergillus niger.
Biochem.Biophys.Res.Commun., 591, 2022
|
|
8IB1
 
 | |
5XOD
 
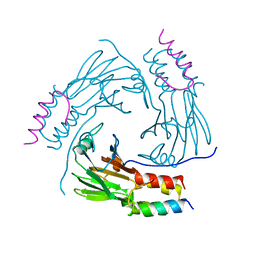 | | Crystal structure of human Smad2-Ski complex | | Descriptor: | Mothers against decapentaplegic homolog 2, Ski oncogene | | Authors: | Miyazono, K, Moriwaki, S, Ito, T, Tanokura, M. | | Deposit date: | 2017-05-27 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Sci Signal, 11, 2018
|
|
5XOC
 
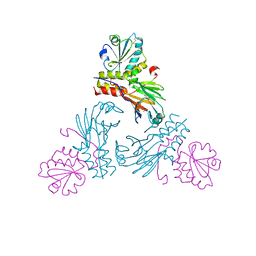 | | Crystal structure of human Smad3-FoxH1 complex | | Descriptor: | Mothers against decapentaplegic homolog 3, Thioredoxin 1,Forkhead box protein H1 | | Authors: | Miyazono, K, Ito, T, Tanokura, M. | | Deposit date: | 2017-05-27 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors
Sci Signal, 11, 2018
|
|
6L2N
 
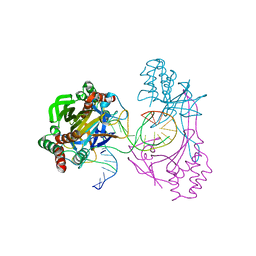 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-3bp-GTAC) complex | | Descriptor: | DNA (5'-D(*TP*CP*AP*GP*CP*AP*GP*TP*AP*CP*TP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*GP*A)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
