4EOP
 
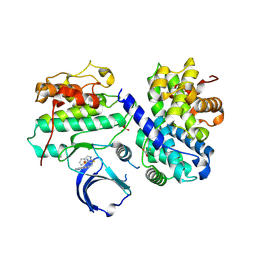 | | Thr 160 phosphorylated CDK2 Q131E - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOO
 
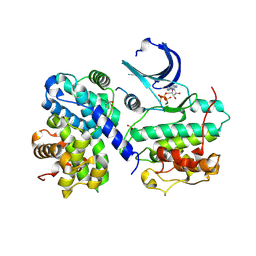 | | Thr 160 phosphorylated CDK2 Q131E - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOJ
 
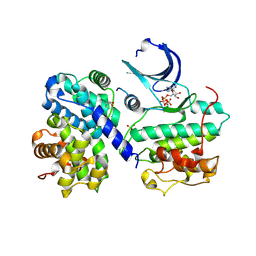 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOK
 
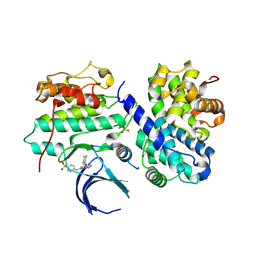 | | Thr 160 phosphorylated CDK2 H84S, Q85M, K89D - human cyclin A3 complex with the inhibitor NU6102 | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, MONOTHIOGLYCEROL, ... | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4EOS
 
 | | Thr 160 phosphorylated CDK2 WT - human cyclin A3 complex with the inhibitor RO3306 | | Descriptor: | (5E)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Cyclin-A2, Cyclin-dependent kinase 2 | | Authors: | Echalier, A, Cot, E, Camasses, A, Hodimont, E, Hoh, F, Sheinerman, F, Krasinska, L, Fisher, D. | | Deposit date: | 2012-04-14 | | Release date: | 2013-02-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | An integrated chemical biology approach provides insight into Cdk2 functional redundancy and inhibitor sensitivity.
Chem.Biol., 19, 2012
|
|
4LHD
 
 | | Crystal structure of Synechocystis sp. PCC 6803 glycine decarboxylase (P-protein), holo form with pyridoxal-5'-phosphate and glycine, closed flexible loop | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, GLYCINE, ... | | Authors: | Hasse, D, Andersson, E, Carlsson, G, Masloboy, A, Hagemann, M, Bauwe, H, Andersson, I. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7959 Å) | | Cite: | Structure of the Homodimeric Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803 Suggests a Mechanism for Redox Regulation.
J.Biol.Chem., 288, 2013
|
|
4LGL
 
 | | Crystal Structure of Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803, apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Glycine dehydrogenase [decarboxylating] | | Authors: | Hasse, D, Andersson, E, Carlsson, G, Masloboy, A, Hagemann, M, Bauwe, H, Andersson, I. | | Deposit date: | 2013-06-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0004 Å) | | Cite: | Structure of the Homodimeric Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803 Suggests a Mechanism for Redox Regulation.
J.Biol.Chem., 288, 2013
|
|
4LHC
 
 | | Crystal structure of Synechocystis sp. PCC 6803 glycine decarboxylase (P-protein), holo form with pyridoxal-5'-phosphate and glycine | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, BICINE, ... | | Authors: | Hasse, D, Andersson, E, Carlsson, G, Masloboy, A, Hagemann, M, Bauwe, H, Andersson, I. | | Deposit date: | 2013-07-01 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structure of the Homodimeric Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803 Suggests a Mechanism for Redox Regulation.
J.Biol.Chem., 288, 2013
|
|
1J1E
 
 | | Crystal structure of the 52kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
3F3A
 
 | | Crystal Structure of LeuT bound to L-Tryptophan and Sodium | | Descriptor: | SODIUM ION, TETRADECANE, TRYPTOPHAN, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
1J1D
 
 | | Crystal structure of the 46kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
3F3D
 
 | | Crystal structure of LeuT bound to L-Methionine and sodium | | Descriptor: | METHIONINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
4D4L
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-chlorophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4D4K
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, 7-(4-methoxyphenyl)-5-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
3F3E
 
 | | Crystal structure of LeuT bound to L-leucine (30 mM) and sodium | | Descriptor: | LEUCINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
4D4M
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, 7-(4-bromophenyl)-5-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
4D4J
 
 | | human PFKFB3 in complex with a pyrrolopyrimidone compound | | Descriptor: | 5-(4-bromophenyl)-7-phenyl-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Stgallay, S.A, Bennett, N, Critchlow, S, Davies, G, Debreczeni, J.E, Evans, N, Holdgate, G, Jones, N.P, Leach, L, Maman, S, McLoughlin, S, Preston, M, Rigoreau, L, Thomas, A, Walker, G, Walsch, J, Ward, R.A, Wheatley, E, Winter, J. | | Deposit date: | 2014-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identifying a Novel Series of Pfkfb3 Inhibitors as a Metabolic Approach to Treating Cancer from Hts, Biophysical and Biochemical Methods
To be Published
|
|
1KU9
 
 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | Descriptor: | hypothetical protein MJ223 | | Authors: | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-01-21 | | Release date: | 2002-12-25 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
3B8Z
 
 | | High Resolution Crystal Structure of the Catalytic Domain of ADAMTS-5 (Aggrecanase-2) | | Descriptor: | CALCIUM ION, N-hydroxy-4-({4-[4-(trifluoromethyl)phenoxy]phenyl}sulfonyl)tetrahydro-2H-pyran-4-carboxamide, ZINC ION, ... | | Authors: | Shieh, H.-S, Williams, J.M, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2).
J.Biol.Chem., 283, 2008
|
|
3F3C
 
 | | Crystal structure of LeuT bound to 4-Fluoro-L-Phenylalanine and sodium | | Descriptor: | 4-FLUORO-L-PHENYLALANINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
1GXW
 
 | | the 2.2 A resolution structure of thermolysin crystallized in presence of potassium thiocyanate | | Descriptor: | CALCIUM ION, LYSINE, THERMOLYSIN, ... | | Authors: | Gaucher, J.F, Selkti, M, Prange, T, Tomas, A. | | Deposit date: | 2002-04-12 | | Release date: | 2002-12-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The 2.2 A Resolution Structure of Thermolysin (Tln) Crystallized in the Presence of Potassium Thiocyanate.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5JH9
 
 | | Crystal structure of prApe1 | | Descriptor: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGE
 
 | |
5JGF
 
 | | Crystal structure of mApe1 | | Descriptor: | Vacuolar aminopeptidase 1, ZINC ION | | Authors: | Noda, N.N, Adachi, W, Inagaki, F. | | Deposit date: | 2016-04-20 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
7JKB
 
 | | 2xVH Fab | | Descriptor: | Anti-Her2, Anti-lysozyme | | Authors: | Lord, D.M, Zhou, Y.F. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Bringing the Heavy Chain to Light: Creating a Symmetric, Bivalent IgG-Like Bispecific.
Antibodies, 9, 2020
|
|
