1A76
 
 | | FLAP ENDONUCLEASE-1 FROM METHANOCOCCUS JANNASCHII | | Descriptor: | FLAP ENDONUCLEASE-1 PROTEIN, MANGANESE (II) ION | | Authors: | Hwang, K.Y, Baek, K, Kim, H, Cho, Y. | | Deposit date: | 1998-03-20 | | Release date: | 1999-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of flap endonuclease-1 from Methanococcus jannaschii.
Nat.Struct.Biol., 5, 1998
|
|
4O6G
 
 | | Rv3902c from M. tuberculosis | | Descriptor: | Uncharacterized protein | | Authors: | Reddy, B.G, Moates, D.B, Kim, H, Green, T.J, Kim, C, Terwilliger, T.J, Delucas, L.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 angstrom resolution X-ray crystal structure of Rv3902c from Mycobacterium tuberculosis.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
1LCP
 
 | |
5XB1
 
 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Ahn, B.J, Jeong, H, Kim, H, Hyun, J, Jung, Y. | | Deposit date: | 2017-03-15 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5YI5
 
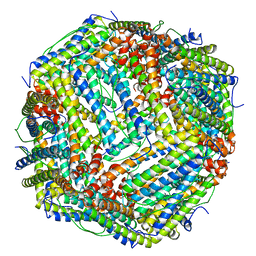 | | human ferritin mutant - E-helix deletion | | Descriptor: | Ferritin heavy chain | | Authors: | Lee, S.G, Yoon, H.R, Ahn, B.J, Jeong, H, Hyun, J, Jung, Y, Kim, H. | | Deposit date: | 2017-10-02 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Four-fold Channel-Nicked Human Ferritin Nanocages for Active Drug Loading and pH-Responsive Drug Release
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3H0T
 
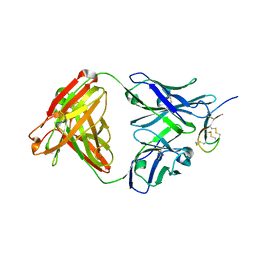 | | Hepcidin-Fab complex | | Descriptor: | Fab fragment, Heavy chain, Light chain, ... | | Authors: | Syed, R, Li, V. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Hepcidin revisited, disulfide connectivity, dynamics, and structure.
J.Biol.Chem., 284, 2009
|
|
4QBS
 
 | |
4QMK
 
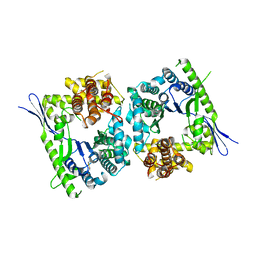 | | Crystal structure of type III effector protein ExoU (exoU) | | Descriptor: | BETA-MERCAPTOETHANOL, Type III secretion system effector protein ExoU | | Authors: | Halavaty, A.S, Tyson, G.H, Zhang, A, Hauser, A.R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-16 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Phosphatidylinositol 4,5-Bisphosphate Binding Domain Mediates Plasma Membrane Localization of ExoU and Other Patatin-like Phospholipases.
J.Biol.Chem., 290, 2015
|
|
6VAT
 
 | |
6VC7
 
 | |
6VDF
 
 | |
4QBQ
 
 | | Crystal structure of DNMT3a ADD domain bound to H3 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone H3, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2014-05-08 | | Release date: | 2015-05-13 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Engineering of a histone-recognition domain in a de novo DNA methyltransferase alters the epigenetic landscape of ESCs
To be Published
|
|
6I60
 
 | | Structure of alpha-L-rhamnosidase from Dictyoglumus thermophilum | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, Alpha-rhamnosidase, TRIETHYLENE GLYCOL | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2018-11-15 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Biochemical Characterization of the alpha-l-RhamnosidaseDtRha fromDictyoglomus thermophilum: Application to the Selective Derhamnosylation of Natural Flavonoids.
Acs Omega, 4, 2019
|
|
8JZM
 
 | | The inhibitor of Toll-like receptor signaling o-vanillin binds covalently to MAL/TIRAP Lys-210 | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Rahaman, M.H, Jia, X, Maxwell, M.J, Mobli, M, Kobe, B. | | Deposit date: | 2023-07-05 | | Release date: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | o-Vanillin binds covalently to MAL/TIRAP Lys-210 but independently inhibits TLR2.
J Enzyme Inhib Med Chem, 39, 2024
|
|
8JPS
 
 | | Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map) | | Descriptor: | Atypical chemokine receptor 1, C-C motif chemokine 7 | | Authors: | Banerjee, R, Khanppnavar, B, Maharana, J, Saha, S, Korkhov, V.M, Shukla, A.K. | | Deposit date: | 2023-06-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism of distinct chemokine engagement and functional divergence of the human Duffy antigen receptor.
Cell, 187, 2024
|
|
5VCH
 
 | |
5VKT
 
 | | Cinnamyl alcohol dehydrogenases (SbCAD4) from Sorghum bicolor (L.) Moench | | Descriptor: | ACETATE ION, Cinnamyl alcohol dehydrogenases (SbCAD4), D-MALATE, ... | | Authors: | Walker, A.M, Jun, S.Y, Kang, C. | | Deposit date: | 2017-04-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | The Enzyme Activity and Substrate Specificity of Two Major Cinnamyl Alcohol Dehydrogenases in Sorghum (Sorghum bicolor), SbCAD2 and SbCAD4.
Plant Physiol., 174, 2017
|
|
4FJO
 
 | | Structure of the Rev1 CTD-Rev3/7-Pol kappa RIR complex | | Descriptor: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | Authors: | Wojtaszek, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Structural basis of Rev1-mediated assembly of a quaternary vertebrate translesion polymerase complex consisting of Rev1, heterodimeric Pol zeta and Pol kappa
J.Biol.Chem., 287, 2012
|
|
5VE8
 
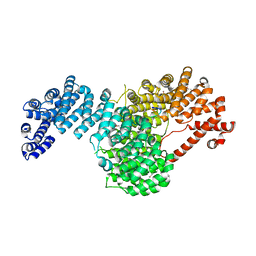 | | Crystal structure of full-length Kluyveromyces lactis Kap123 with histone H3 1-28 | | Descriptor: | Histone H3, Kap123 | | Authors: | An, S, Yoon, J, Song, J.-J, Cho, U.-S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based nuclear import mechanism of histones H3 and H4 mediated by Kap123.
Elife, 6, 2017
|
|
4EN1
 
 | |
2LM4
 
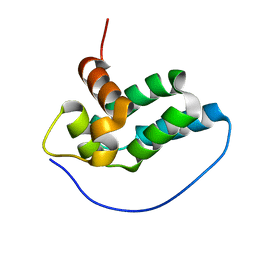 | | Solution NMR Structure of mitochondrial succinate dehydrogenase assembly factor 2 from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium Target YT682A | | Descriptor: | Succinate dehydrogenase assembly factor 2, mitochondrial | | Authors: | Eletsky, A, Winge, D.R, Lee, H, Lee, D, Kohan, E, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-11-22 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast succinate dehydrogenase flavinylation factor sdh5 reveals a putative sdh1 binding site.
Biochemistry, 51, 2012
|
|
2LYQ
 
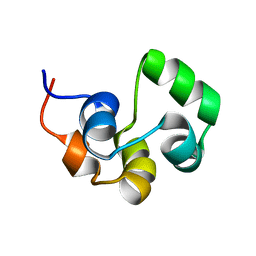 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
7CM0
 
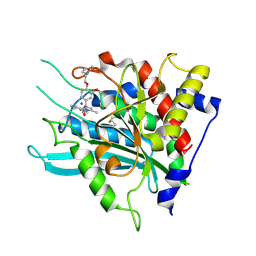 | | Crystal structure of a glutaminyl cyclase in complex with NHV-1009 | | Descriptor: | 1-(cyclopentylmethyl)-1-[3-methoxy-4-(2-morpholin-4-ylethoxy)phenyl]-3-[3-(5-methylimidazol-1-yl)propyl]urea, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Lee, J.W, Song, J.Y, Jang, T.H, Ha, J.H. | | Deposit date: | 2020-07-23 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of highly potent human glutaminyl cyclase (QC) inhibitors as anti-Alzheimer's agents by the combination of pharmacophore-based and structure-based design.
Eur.J.Med.Chem., 226, 2021
|
|
2LYR
 
 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
