4XWT
 
 | | Crystal structure of RNase J complexed with UMP | | Descriptor: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | Deposit date: | 2015-01-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
5BNM
 
 | | E. coli FabH with Small Molecule Inhibitor 1 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3, N-{[3'-(hydroxymethyl)biphenyl-4-yl]methyl}benzenesulfonamide, SULFATE ION, ... | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
4Z8D
 
 | |
5BNS
 
 | | E. coli Fabh with small molecule inhibitor 2 | | Descriptor: | 1-{5-[2-fluoro-5-(hydroxymethyl)phenyl]pyridin-2-yl}-N-(quinolin-6-ylmethyl)piperidine-4-carboxamide, 3-oxoacyl-[acyl-carrier-protein] synthase 3 | | Authors: | Kazmirski, S.L, McKinney, D.C. | | Deposit date: | 2015-05-26 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibacterial FabH Inhibitors with Mode of Action Validated in Haemophilus influenzae by in Vitro Resistance Mutation Mapping.
Acs Infect Dis., 2, 2016
|
|
7E2U
 
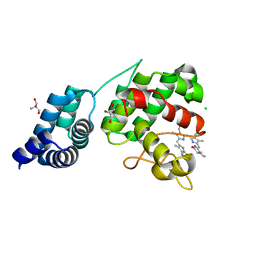 | | Synechocystis GUN4 in complex with phytochrome | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[5-[(~{Z})-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, ... | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2T
 
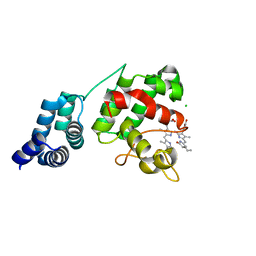 | | Synechocystis GUN4 in complex with phycocyanobilin | | Descriptor: | 3-[2-[(~{Z})-[5-[(~{Z})-[(4~{R})-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(~{Z})-(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, CHLORIDE ION, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7E2S
 
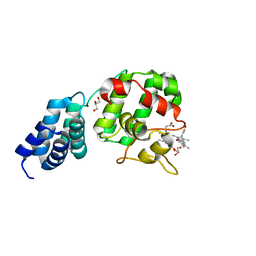 | | Synechocystis GUN4 in complex with biliverdin IXa | | Descriptor: | BILIVERDINE IX ALPHA, GLYCEROL, Ycf53-like protein | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
7YTJ
 
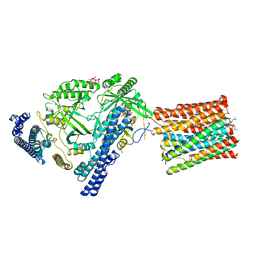 | | Cryo-EM structure of VTC complex | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, INOSITOL HEXAKISPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Guan, Z.Y, Chen, J, Liu, R.W, Chen, Y.K, Xing, Q, Du, Z.M, Liu, Z. | | Deposit date: | 2022-08-15 | | Release date: | 2023-02-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The cytoplasmic synthesis and coupled membrane translocation of eukaryotic polyphosphate by signal-activated VTC complex.
Nat Commun, 14, 2023
|
|
4P73
 
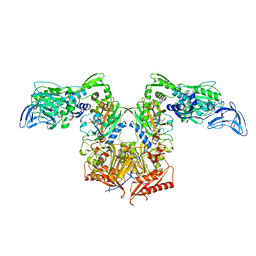 | | PheRS in complex with compound 1a | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P75
 
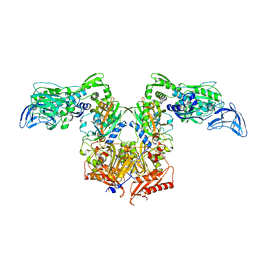 | | PheRS in complex with compound 4a | | Descriptor: | 3-(3-methoxyphenyl)-5-(trifluoromethyl)-1H-pyrazole, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P72
 
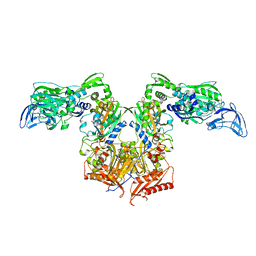 | | PheRS in complex with compound 2a | | Descriptor: | 2-{3-[(4-chloropyridin-2-yl)amino]phenoxy}-N-methylacetamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
7X31
 
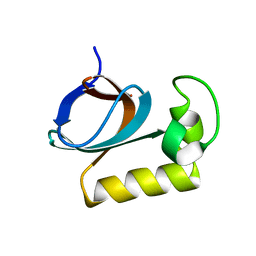 | | solution structure of an anti-CRISPR protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1) | | Authors: | Zhao, Y, Yang, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
4P74
 
 | | PheRS in complex with compound 3a | | Descriptor: | N-[(3S)-1,1-dioxidotetrahydrothiophen-3-yl]-2-[(4-methylphenoxy)methyl]-1,3-thiazole-4-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P71
 
 | | Apo PheRS from P. aeuriginosa | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
3QNU
 
 | |
7D6H
 
 | |
7E35
 
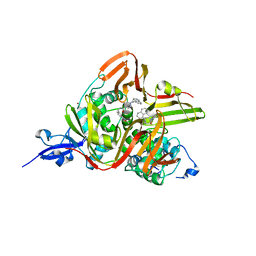 | | Crystal structure of the SARS-CoV-2 papain-like protease (PLPro) C112S mutant bound to compound S43 | | Descriptor: | N-[(3-acetamidophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Non-structural protein 3, ZINC ION | | Authors: | Liu, J, Wang, Y, Xu, X, Pan, L. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2.
Cell Chem Biol, 28, 2021
|
|
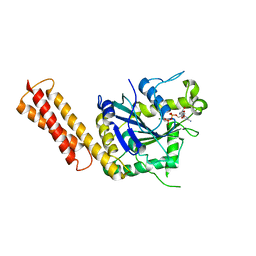
3QOF
 
 | |
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
7E1R
 
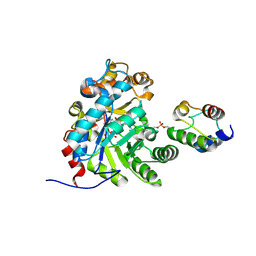 | | Crystal structure of Dehydrogenase/isomerase FabX from Helicobacter pylori in complex with holo-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.795 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1Q
 
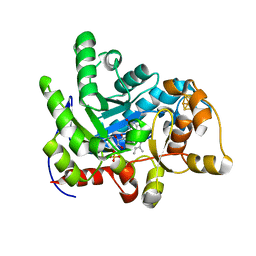 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori | | Descriptor: | 2-nitropropane dioxygenase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7E1S
 
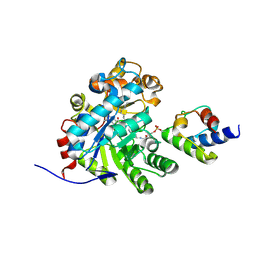 | | Crystal structure of dehydrogenase/isomerase FabX from Helicobacter pylori in complex with octanoyl-ACP | | Descriptor: | 2-nitropropane dioxygenase, Acyl carrier protein,Acyl carrier protein, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Zhou, J.S, Zhang, L, Zhang, L. | | Deposit date: | 2021-02-03 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Helicobacter pylori FabX contains a [4Fe-4S] cluster essential for unsaturated fatty acid synthesis.
Nat Commun, 12, 2021
|
|
7Y5K
 
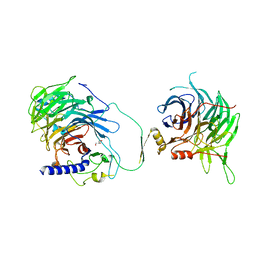 | | Crystal structure of human CAF-1 core complex in spacegroup C2221 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y61
 
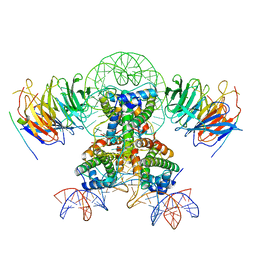 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5O
 
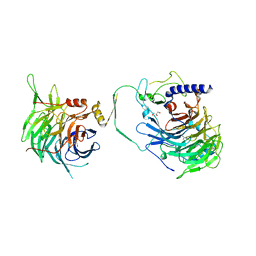 | | Crystal structure of human CAF-1 core complex in spacegroup P21 | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, GLYCEROL, ... | | Authors: | Liu, C.P, Wang, M.Z, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
