6I1X
 
 | | Aeromonas hydrophila ExeD | | Descriptor: | Type II secretion system protein D | | Authors: | Contreras-Martel, C, Farias Estrozi, L. | | Deposit date: | 2018-10-30 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system.
Plos Pathog., 15, 2019
|
|
1FTW
 
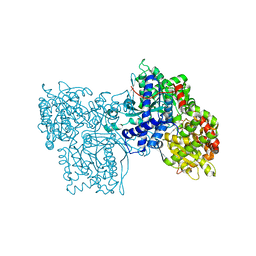 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3,8,9,10-tetrahydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
6R72
 
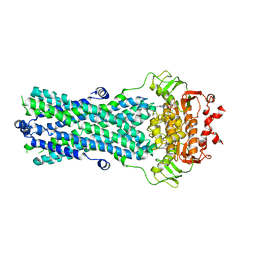 | | Crystal structure of BmrA-E504A in an outward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Multidrug exporter ATP-binding cassette | | Authors: | Chaptal, V, Zampieri, V, Kilburg, A, Magnard, S, Falson, P. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
6R81
 
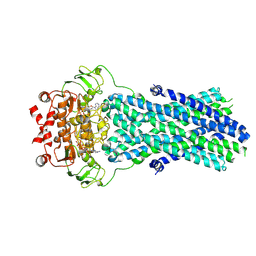 | | Multidrug resistance transporter BmrA mutant E504A bound with ATP and Mg solved by Cryo-EM | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION | | Authors: | Wiseman, B, Chaptal, V, Zampieri, V, Magnard, S, Hogbom, M, Falson, P. | | Deposit date: | 2019-03-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Substrate-bound and substrate-free outward-facing structures of a multidrug ABC exporter.
Sci Adv, 8, 2022
|
|
1FTQ
 
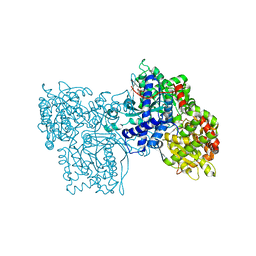 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | (5S,7R,8S,9S,10R)-3-amino-8,9,10-trihydroxy-7-(hydroxymethyl)-6-oxa-1,3-diazaspiro[4.5]decane-2,4-dione, GLYCOGEN PHOSPHORYLASE, INOSINIC ACID, ... | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
5N2U
 
 | | Influenza D virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Labaronne, A, Ruigrok, R.W.H, Crepin, T. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the nucleoprotein of Influenza D shows that all Orthomyxoviridae nucleoproteins have a similar NPCORE, with or without a NPTAILfor nuclear transport.
Sci Rep, 9, 2019
|
|
6S44
 
 | |
1FS4
 
 | | Structures of glycogen phosphorylase-inhibitor complexes and the implications for structure-based drug design | | Descriptor: | 1-DEOXY-1-METHOXYCARBAMIDO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-08 | | Release date: | 2000-10-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
2QRQ
 
 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-methylphenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(4-methylphenyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
1FU4
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | GLYCOGEN PHOSPHORYLASE, N-[(5S,7R,8S,9S,10R)-8,9,10-trihydroxy-7-(hydroxymethyl)-2,4-dioxo-6-oxa-1,3-diazaspiro[4.5]dec-3-yl]acetamide, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
2QRM
 
 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-nitrophenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(4-nitrophenyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, Glycogen phosphorylase, muscle form | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
1FU7
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | GLYCOGEN PHOSPHORYLASE, N-(methoxycarbonyl)-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1FTY
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 8,9,10-TRIHYDROXY-7-HYDROXYMETHYL-3-METHYL-6-OXA-1,3-DIAZA-SPIRO[4.5]DECANE-2,4-DIONE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-13 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
1OCY
 
 | | Structure of the receptor-binding domain of the bacteriophage T4 short tail fibre | | Descriptor: | BACTERIOPHAGE T4 SHORT TAIL FIBRE, CITRIC ACID, SULFATE ION, ... | | Authors: | Thomassen, E, Gielen, G, Schuetz, M, Miller, S, van Raaij, M.J. | | Deposit date: | 2003-02-11 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Structure of the Receptor-Binding Domain of the Bacteriophage T4 Short Tail Fibre Reveals a Knitted Trimeric Metal-Binding Fold
J.Mol.Biol., 331, 2003
|
|
2QRP
 
 | | Glycogen Phosphorylase b in complex with (1R)-3'-(2-naphthyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(2-naphthyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
1FU8
 
 | | STRUCTURES OF GLYCOGEN PHOSPHORYLASE-INHIBITOR COMPLEXES AND THE IMPLICATIONS FOR STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 1-DEOXY-1-ACETYLAMINO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, GLYCOGEN PHOSPHORYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Watson, K.A, Tsitsanou, K.E, Gregoriou, M, Zographos, S.E, Skamnaki, V.T, Oikonomakos, N.G, Fleet, G.W, Johnson, L.N. | | Deposit date: | 2000-09-14 | | Release date: | 2000-10-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Kinetic and crystallographic studies of glucopyranose spirohydantoin and glucopyranosylamine analogs inhibitors of glycogen phosphorylase.
Proteins, 61, 2005
|
|
2QRG
 
 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-methoxyphenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (5R,7R,8S,9S,10R)-7-(HYDROXYMETHYL)-3-(4-METHOXYPHENYL)-1,6-DIOXA-2-AZASPIRO[4.5]DEC-2-ENE-8,9,10-TRIOL, Glycogen phosphorylase, muscle form | | Authors: | Kizilis, G, Alexacou, K.-M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
2QRH
 
 | | Glycogen Phosphorylase b in complex with (1R)-3'-phenylspiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-phenyl-1,6-dioxa-2-azaspiro[4.5]dec-2-ene-8,9,10-triol, Glycogen phosphorylase, muscle form | | Authors: | Kizilis, G, Alexacou, K.-M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
7PCD
 
 | | HER2 IN COMPLEX WITH A COVALENT INHIBITOR | | Descriptor: | 1-[4-[4-[[3,5-bis(chloranyl)-4-([1,2,4]triazolo[1,5-a]pyridin-7-yloxy)phenyl]amino]pyrimido[5,4-d]pyrimidin-6-yl]piperazin-1-yl]-4-(3-fluoranylazetidin-1-yl)butan-1-one, Receptor tyrosine-protein kinase erbB-2 | | Authors: | Bader, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery of potent and selective HER2 inhibitors with efficacy against HER2 exon 20 insertion-driven tumors, which preserve wild-type EGFR signaling.
Nat Cancer, 3, 2022
|
|
7XXN
 
 | | HapR Quadruple mutant, bound to Qstatin | | Descriptor: | 1-(5-bromanylthiophen-2-yl)sulfonylpyrazole, GLYCEROL, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XY0
 
 | | HapR Double mutant Y76F, F171C | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-31 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7Y4J
 
 | | HapR_Triple mutant Y76F, L97I, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXS
 
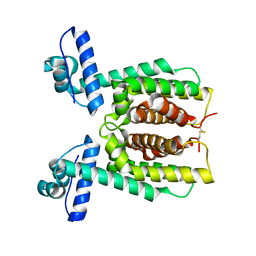 | | HapR mutant I141V | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XYI
 
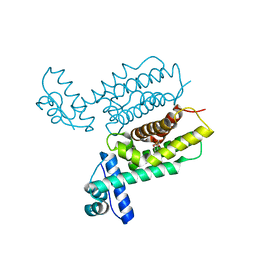 | | HapR Quadruple mutant Y76F, L97I, I141V, F171C | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
7XXO
 
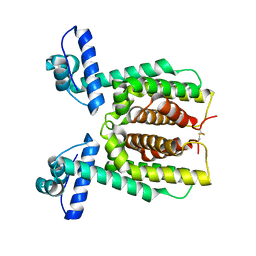 | | HapR Native in CHES buffer pH 9.5 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Basu Choudhury, G, Chaudhari, V, Ray Chaudhuri, S, Datta, S. | | Deposit date: | 2022-05-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Diversity in the ligand binding pocket of HapR attributes to its uniqueness towards several inhibitors with respect to other homologues - A structural and molecular perspective.
Int.J.Biol.Macromol., 233, 2023
|
|
