8BTJ
 
 | | Murine cytomegalovirus protein M35 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MALONATE ION, Protein M35 | | Authors: | Schmelz, S, Van den Heuvel, J, Blankenfeldt, W. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Cytomegalovirus M35 Protein Directly Binds to the Interferon-beta Enhancer and Modulates Transcription of Ifnb1 and Other IRF3-Driven Genes.
J.Virol., 97, 2023
|
|
7B3O
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
6O16
 
 | | Crystal structure of murine DHX37 in complex with RNA | | Descriptor: | DEAH (Asp-Glu-Ala-His) box polypeptide 37, RNA (5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Boneberg, F, Brandmann, T, Kobel, L, van den Heuvel, J, Bargsten, K, Bammert, L, Kutay, U, Jinek, M. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.875 Å) | | Cite: | Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis.
Rna, 25, 2019
|
|
6ZXE
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State F2 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZXF
 
 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State G | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZXD
 
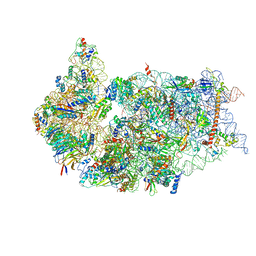 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State F1 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZXH
 
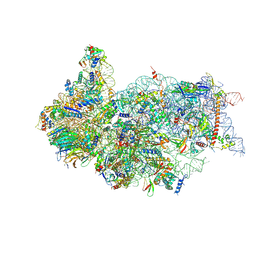 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H2 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
6ZXG
 
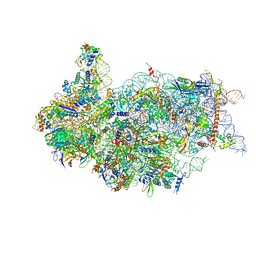 | | Cryo-EM structure of a late human pre-40S ribosomal subunit - State H1 | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Ameismeier, M, Zemp, I, van den Heuvel, J, Thoms, M, Berninghausen, O, Kutay, U, Beckmann, R. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the final steps of human 40S ribosome maturation.
Nature, 587, 2020
|
|
5LSP
 
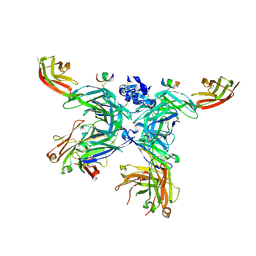 | | 107_A07 Fab in complex with fragment of the Met receptor | | Descriptor: | 107_A07 Fab heavy chain, 107_A07 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DiCara, D, Chirgadze, D.Y, Pope, A, Karatt-Vellatt, A, Winter, A, van den Heuvel, J, Gherardi, E, McCafferty, J. | | Deposit date: | 2016-09-05 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.605 Å) | | Cite: | Characterization and structural determination of a new anti-MET function-blocking antibody with binding epitope distinct from the ligand binding domain.
Sci Rep, 7, 2017
|
|
2W0R
 
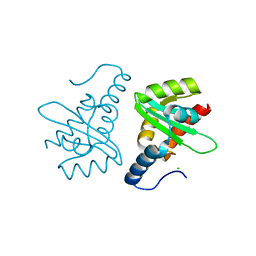 | | Crystal structure of the mutated N263D YscU C-terminal domain | | Descriptor: | CHLORIDE ION, YSCU | | Authors: | Wiesand, U, Sorg, I, Amstutz, M, Wagner, S, Van Den Heuvel, J, Luehrs, T, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the Type III Secretion Recognition Protein Yscu from Yersinia Enterocolitica
J.Mol.Biol., 385, 2009
|
|
2UZX
 
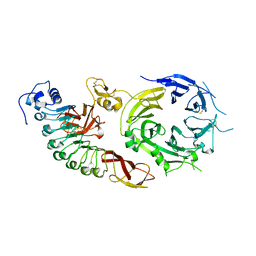 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein InlB: Crystal form I | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, Van Den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
2V5G
 
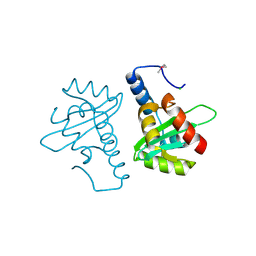 | | Crystal structure of the mutated N263A YscU C-terminal domain | | Descriptor: | CHLORIDE ION, YSCU | | Authors: | Wiesand, U, Sorg, I, Amstutz, M, Wagner, S, Van Den Heuvel, J, Luehrs, T, Cornelis, G.R, Heinz, D.W. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-04 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Type III Secretion Recognition Protein Yscu from Yersinia Enterocolitica
J.Mol.Biol., 385, 2009
|
|
2UZY
 
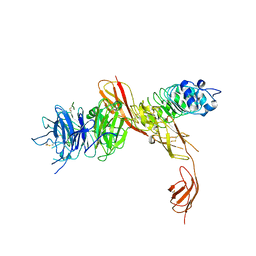 | | Structure of the human receptor tyrosine kinase Met in complex with the Listeria monocytogenes invasion protein inlb: low resolution, Crystal form II | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR, INTERNALIN B | | Authors: | Niemann, H.H, Jager, V, Butler, P.J.G, van den Heuvel, J, Schmidt, S, Ferraris, D, Gherardi, E, Heinz, D.W. | | Deposit date: | 2007-05-02 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Human Receptor Tyrosine Kinase met in Complex with the Listeria Invasion Protein Inlb
Cell(Cambridge,Mass.), 130, 2007
|
|
5IV3
 
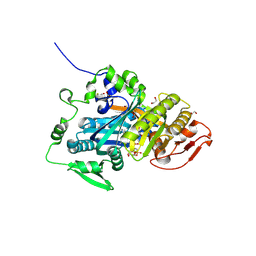 | | Crystal structure of human soluble adenylyl cyclase in complex with alpha,beta-methyleneadenosine-5'-triphosphate and the allosteric inhibitor LRE1 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-N~4~-cyclopropyl-N~4~-[(thiophen-2-yl)methyl]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of LRE1 as a specific and allosteric inhibitor of soluble adenylyl cyclase.
Nat.Chem.Biol., 12, 2016
|
|
4UST
 
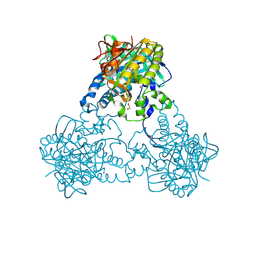 | |
4USV
 
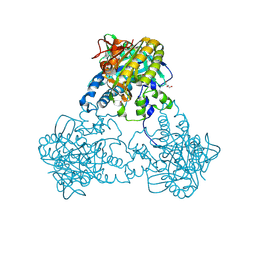 | |
4USW
 
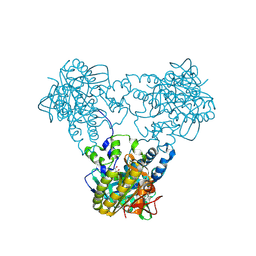 | | Crystal structure of human soluble Adenylyl Cyclase with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2014-07-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of Human Soluble Adenylyl Cyclase and Crystal Structures of its Nucleotide Complexes -Implications for Cyclase Catalysis and Evolution.
FEBS J., 281, 2014
|
|
6R6U
 
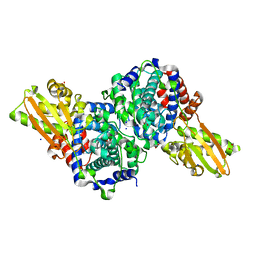 | | Crystal structure of human cis-aconitate decarboxylase | | Descriptor: | (R,R)-2,3-BUTANEDIOL, Cis-aconitate decarboxylase, SODIUM ION | | Authors: | Lukat, P, Chen, F, Saile, K, Buessow, K, Pessler, F, Blankenfeldt, W. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Crystal structure ofcis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2OMY
 
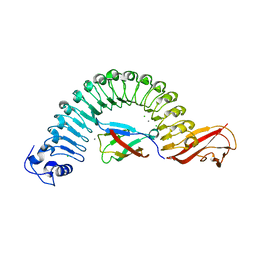 | | Crystal structure of InlA S192N/hEC1 complex | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epithelial-cadherin, ... | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
2OMW
 
 | | Crystal structure of InlA S192N Y369S/mEC1 complex | | Descriptor: | CHLORIDE ION, Epithelial-cadherin, Internalin-A | | Authors: | Wollert, T, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2007-01-23 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extending the host range of Listeria monocytogenes by rational protein design.
Cell(Cambridge,Mass.), 129, 2007
|
|
8CNH
 
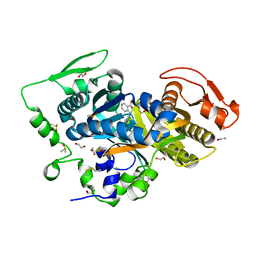 | |
8COJ
 
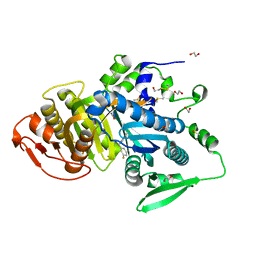 | |
4CLL
 
 | | Crystal structure of human soluble Adenylyl Cyclase in complex with bicarbonate | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE CYCLASE TYPE 10, BICARBONATE ION, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CLU
 
 | | Crystal structure of human soluble Adenylyl Cyclase with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CLY
 
 | | Crystal structure of human soluble Adenylyl Cyclase soaked with biselenite | | Descriptor: | 1,2-ETHANEDIOL, ADENYLATE CYCLASE TYPE 10, BISELENITE ION, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
