1ZOS
 
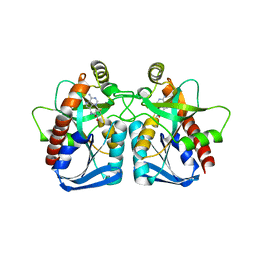 | | Structure of 5'-methylthionadenosine/S-Adenosylhomocysteine nucleosidase from S. pneumoniae with a transition-state inhibitor MT-ImmA | | Descriptor: | (3S,4R)-2-(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)-5-[(METHYLSULFANYL)METHYL]PYRROLIDINE-3,4-DIOL, 5'-methylthioadenosine / S-adenosylhomocysteine nucleosidase | | Authors: | Shi, W, Singh, V, Zhen, R, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2005-05-13 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and inhibition of a quorum sensing target from Streptococcus pneumoniae.
Biochemistry, 45, 2006
|
|
7X8L
 
 | |
1RR2
 
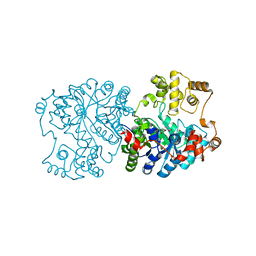 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to 2-ketobutyric acid | | Descriptor: | 2-KETOBUTYRIC ACID, COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-08 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1S3H
 
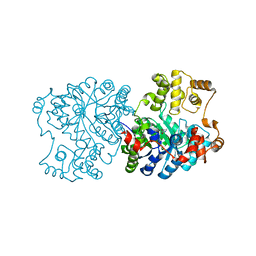 | | Propionibacterium shermanii transcarboxylase 5S subunit A59T | | Descriptor: | COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2004-01-13 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1RQE
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to oxaloacetate | | Descriptor: | COBALT (II) ION, OXALOACETATE ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-05 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1RQH
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit bound to pyruvic acid | | Descriptor: | COBALT (II) ION, PYRUVIC ACID, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-05 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1RQB
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit | | Descriptor: | COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1U5J
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit, Met186Ile | | Descriptor: | COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2004-07-27 | | Release date: | 2004-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1P9L
 
 | | Structure of M. tuberculosis dihydrodipicolinate reductase in complex with NADH and 2,6 PDC | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PYRIDINE-2,6-DICARBOXYLIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes. Structural and mutagenic analysis of relaxed nucleotide specificity.
Biochemistry, 42, 2003
|
|
1ARZ
 
 | | ESCHERICHIA COLI DIHYDRODIPICOLINATE REDUCTASE IN COMPLEX WITH NADH AND 2,6 PYRIDINE DICARBOXYLATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE, PHOSPHATE ION, ... | | Authors: | Scapin, G, Reddy, S.G, Zheng, R, Blanchard, J.S. | | Deposit date: | 1997-08-08 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of Escherichia coli dihydrodipicolinate reductase in complex with NADH and the inhibitor 2,6-pyridinedicarboxylate.
Biochemistry, 36, 1997
|
|
1BWZ
 
 | |
1C3V
 
 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
6KF1
 
 | | Microbial Hormone-sensitive lipase- E53 mutant S162A | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HEXANOIC ACID, ... | | Authors: | Xiaochen, Y, Zhengyang, L, Xuewei, X, Jixi, L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Functional and Structural Insights into Environmental Adaptation of a Novel Hormone-sensitive Lipase, E53, Obtained from Erythrobacter longus
To Be Published
|
|
6KF5
 
 | | Microbial Hormone-sensitive lipase E53 mutant I256L | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X, Li, Z.Y, Li, J, Xu, X.W. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 mutant I256L
To Be Published
|
|
6KEU
 
 | | Wildtype E53, a microbial HSL esterase | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X.C, Li, Z.Y, Xu, X.W, Li, J.X. | | Deposit date: | 2019-07-05 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Wildtype E53, a microbial HSL esterase
To Be Published
|
|
1WJF
 
 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
6LDZ
 
 | | Crystal structure of Rv0222 from Mycobacterium tuberculosis | | Descriptor: | Probable enoyl-CoA hydratase EchA1 (Enoyl hydrase) (Unsaturated acyl-CoA hydratase) (Crotonase) | | Authors: | Li, J, Ran, Y.J, Wang, L, Wu, J.H, Ge, B.X, Rao, Z.H. | | Deposit date: | 2019-11-23 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host-mediated ubiquitination of a mycobacterial protein suppresses immunity.
Nature, 577, 2020
|
|
4TN2
 
 | | NS5b in complex with lactam-thiophene carboxylic acids | | Descriptor: | 3-[(2R)-2-cyclohexyl-5-oxopyrrolidin-1-yl]-5-phenylthiophene-2-carboxylic acid, Genome polyprotein | | Authors: | Chopra, R. | | Deposit date: | 2014-06-02 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and synthesis of lactam-thiophene carboxylic acids as potent hepatitis C virus polymerase inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4TLR
 
 | | NS5b in complex with lactam-thiophene carboxylic acids | | Descriptor: | 3-{(2R,5R)-5-cyclohexyl-2-[(2R)-2-hydroxypropyl]-3-oxomorpholin-4-yl}-5-(3,3-dimethylbut-1-yn-1-yl)thiophene-2-carboxylic acid, 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, NS5b | | Authors: | Chopra, R. | | Deposit date: | 2014-05-30 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Design and synthesis of lactam-thiophene carboxylic acids as potent hepatitis C virus polymerase inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2EZX
 
 | |
2EZY
 
 | |
7SSB
 
 | | Co-structure of PKG1 regulatory domain with compound 33 | | Descriptor: | 4-({(2S,3S)-3-[(1S)-1-(3,5-dichlorophenyl)-2-hydroxyethoxy]-2-phenylpiperidin-1-yl}methyl)-3-nitrobenzoic acid, cGMP-dependent protein kinase 1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2021-11-10 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization and Mechanistic Investigations of Novel Allosteric Activators of PKG1 alpha.
J.Med.Chem., 65, 2022
|
|
1QCK
 
 | |
2EZZ
 
 | |
1WJB
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
