1JXC
 
 | | Minimized NMR structure of ATT, an Arabidopsis trypsin/chymotrypsin inhibitor | | Descriptor: | Putative trypsin inhibitor ATTI-2 | | Authors: | Zhao, Q, Chae, Y.K, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2001-09-06 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of ATTp, an Arabidopsis thaliana Trypsin Inhibitor
Biochemistry, 41, 2002
|
|
4A4R
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GMP*GP*AP*CP*CP*CP*GP*GP*CP*UP*AP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4T
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*UP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4S
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*CP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4A4U
 
 | | UNAC Tetraloops: To What Extent Can They Mimic GNRA Tetraloops | | Descriptor: | 5'-R(*GP*GP*AP*CP*CP*CP*GP*GP*CP*UP*GP*AP*CP*GP *CP*UP*GP*GP*GP*UP*CP*C)-3' | | Authors: | Zhao, Q, Huang, H, Nagaswamy, U, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2011-10-20 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unac Tetraloops: To What Extent Can They Mimic Gnra Tetraloops
Biopolymers, 97, 2012
|
|
4Z7F
 
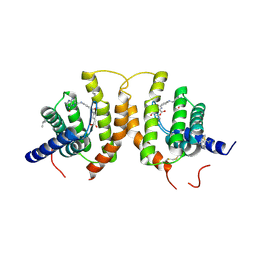 | | Crystal structure of FolT bound with folic acid | | Descriptor: | FOLIC ACID, Folate ECF transporter | | Authors: | Zhao, Q, Wang, C.C, Wang, C.Y, Zhang, P. | | Deposit date: | 2015-04-07 | | Release date: | 2015-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters
Nat Commun, 6, 2015
|
|
4REB
 
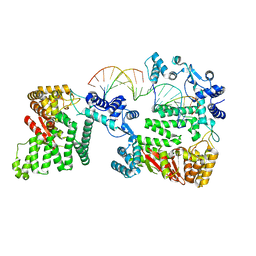 | | Structural Insights into 5' Flap DNA Unwinding and Incision by the Human FAN1 Dimer | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
4REC
 
 | | A nuclease-DNA complex form 3 | | Descriptor: | DNA (40-MER), Fanconi-associated nuclease 1, IODIDE ION | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
4REA
 
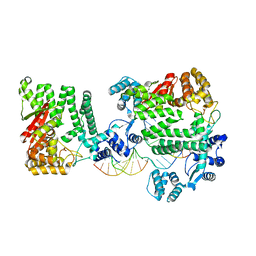 | | A Nuclease DNA complex | | Descriptor: | DNA (5'-D(*TP*GP*CP*TP*CP*GP*CP*CP*AP*C)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
3D23
 
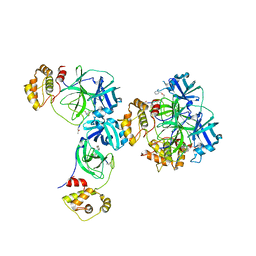 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
1YDU
 
 | | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain | | Descriptor: | At5g01610 | | Authors: | Zhao, Q, Cornilescu, C.C, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-26 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain
To be Published
|
|
4NE6
 
 | | Human MHF1-MHF2 complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NE5
 
 | | Human MHF1-MHF2 complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NE1
 
 | | Human MHF1 MHF2 DNA complexes | | Descriptor: | Centromere protein S, Centromere protein X, DNA (26-MER) | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NE3
 
 | | Human MHF1-MHF2 complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2013-12-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8007 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NDY
 
 | | Human MHF1-MHF2 DNA complex | | Descriptor: | Centromere protein S, Centromere protein X, DNA (26-MER) | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.999 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
1B1C
 
 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
2PN3
 
 | | Crystal Structure of Hepatitis C Virus IRES Subdomain IIa | | Descriptor: | 5'-R(*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*AP*CP*GP*CP*C)-3', 5'-R(*GP*CP*GP*(5BU)P*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*(5BU)P*CP*CP*GP*G)-3', MAGNESIUM ION | | Authors: | Zhao, Q, Han, Q, Kissinger, C.R, Hermann, T, Thompson, P.A. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of hepatitis C virus IRES subdomain IIa.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2PN4
 
 | | Crystal Structure of Hepatitis C Virus IRES Subdomain IIa | | Descriptor: | 5'-R(*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*AP*CP*GP*CP*C)-3', 5'-R(*GP*CP*GP*(5BU)P*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*(5BU)P*CP*CP*GP*G)-3', STRONTIUM ION | | Authors: | Zhao, Q, Han, Q, Kissinger, C.R, Thompson, P.A. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of hepatitis C virus IRES subdomain IIa.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1A6Y
 
 | | REVERBA ORPHAN NUCLEAR RECEPTOR/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*(5IT)P*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3'), ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Zhao, Q, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 1998-03-04 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elements of an orphan nuclear receptor-DNA complex.
Mol.Cell, 1, 1998
|
|
2HEM
 
 | | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit. | | Descriptor: | 5'-R(P*GP*GP*GP*AP*AP*GP*GP*CP*GP*CP*UP*UP*CP*GP*GP*CP*GP*UP*CP*GP*GP*CP*CP*C)-3' | | Authors: | Zhao, Q, Nagaswamy, U, Lee, H, Xia, Y, Gao, X, Fox, G. | | Deposit date: | 2006-06-21 | | Release date: | 2006-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and Mg2+ binding of an RNA segment that underlies the L7/L12 stalk in the E.coli 50S ribosomal subunit
Nucleic Acids Res., 33, 2005
|
|
1BY4
 
 | | STRUCTURE AND MECHANISM OF THE HOMODIMERIC ASSEMBLY OF THE RXR ON DNA | | Descriptor: | DNA (5'-D(*C*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*A)-3'), PROTEIN (RETINOIC ACID RECEPTOR RXR-ALPHA), ... | | Authors: | Zhao, Q, Chasse, S.A, Devarakonda, S, Sierk, M.L, Ahvazi, B, Sigler, P.B, Rastinejad, F. | | Deposit date: | 1998-10-22 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of RXR-DNA interactions.
J.Mol.Biol., 296, 2000
|
|
2NS2
 
 | | Crystal Structure of Spindlin1 | | Descriptor: | PHOSPHATE ION, Spindlin-1 | | Authors: | Zhao, Q, Qin, L, Jiang, F, Wu, B, Yue, W, Xu, F, Rong, Z, Yuan, H, Xie, X, Gao, Y, Bai, C, Bartlam, M. | | Deposit date: | 2006-11-02 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human spindlin1. Tandem tudor-like domains for cell cycle regulation
J.Biol.Chem., 282, 2007
|
|
8K8K
 
 | | Structure of Klebsiella pneumonia ModA | | Descriptor: | Molybdate transporter periplasmic protein | | Authors: | Zhao, Q, Bartlam, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural analysis of molybdate binding protein ModA from Klebsiella pneumoniae.
Biochem.Biophys.Res.Commun., 681, 2023
|
|
8K8L
 
 | |
