4BAC
 
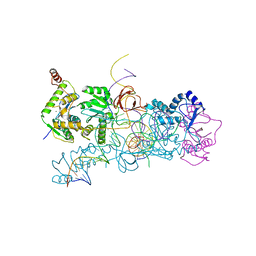 | | prototype foamy virus strand transfer complexes on product DNA | | Descriptor: | 5'-D(*AP*GP*GP*AP*GP*CP*CP*AP*AP*GP*AP*CP*GP*GP *AP*TP*CP)-3', 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*TP*GP*TP*AP)-3', DNA (38-MER), ... | | Authors: | Yin, Z, Lapkouski, M, Yang, W, Craigie, R. | | Deposit date: | 2012-09-12 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Assembly of Prototype Foamy Virus Strand Transfer Complexes on Product DNA Bypassing Catalysis of Integration.
Protein Sci., 21, 2012
|
|
4K3O
 
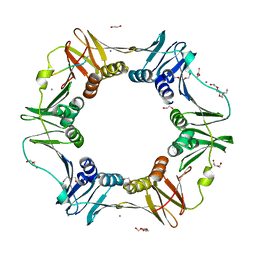 | | E. coli sliding clamp in complex with AcQADLF | | Descriptor: | (ACE)QADLF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3K
 
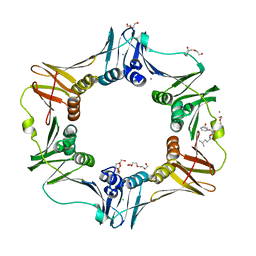 | |
4K3P
 
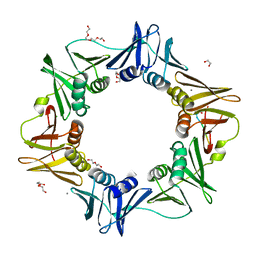 | | E. coli sliding clamp in complex with AcQLALF | | Descriptor: | (ACE)QLALF, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3R
 
 | | E. coli sliding clamp in complex with AcQLDLA | | Descriptor: | (ACE)QLDLA, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3S
 
 | | E. coli sliding clamp in P1 crystal space group | | Descriptor: | CALCIUM ION, DNA polymerase III subunit beta, O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-11 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
4K3L
 
 | | E. coli sliding clamp in complex with AcLF dipeptide | | Descriptor: | 1,2-ETHANEDIOL, ACETYL GROUP, CALCIUM ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Thermodynamic Dissection of Linear Motif Recognition by the E. coli Sliding Clamp
J.Med.Chem., 56, 2013
|
|
7AK6
 
 | | Cryo-EM structure of ND6-P25L mutant respiratory complex I from Mus musculus at 3.8 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
7AK5
 
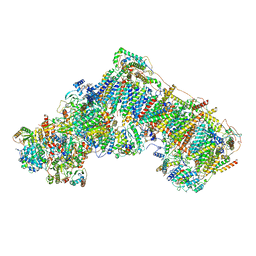 | | Cryo-EM structure of respiratory complex I in the deactive state from Mus musculus at 3.2 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
6UCM
 
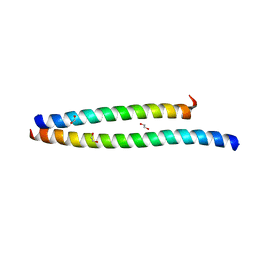 | | Transcription factor DeltaFosB bZIP domain self-assembly, type-II crystal | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
6UCL
 
 | |
6UCI
 
 | | Transcription factor DeltaFosB bZIP domain self-assembly, oxidized form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein fosB | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2019-09-16 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Self-assembly of the bZIP transcription factor Delta FosB.
Curr Res Struct Biol, 2, 2020
|
|
8QJ7
 
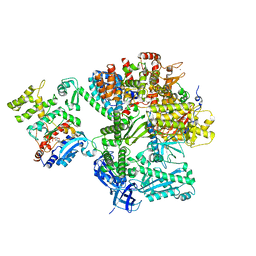 | |
6P19
 
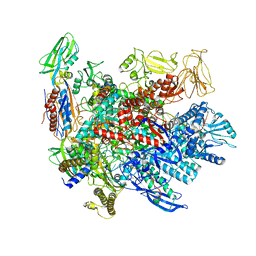 | | Q21 transcription antitermination complex: loaded complex | | Descriptor: | DNA (123-MER) fragment carrying phage-21 pR' promoter, pause element, and transcribed region, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P18
 
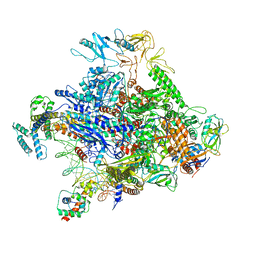 | | Q21 transcription antitermination complex: loading complex | | Descriptor: | DNA (67-MER) fragment carrying phage-21 pR' promoter and pause element, nontemplate strand, template strand, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6P1B
 
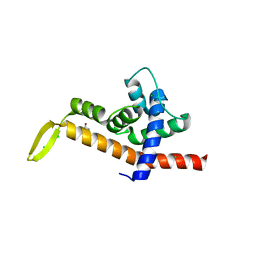 | | Transcription antitermination factor Q21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Q protein | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
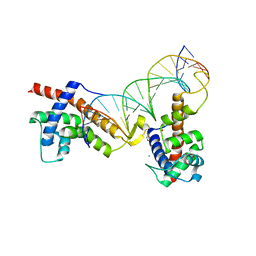
6P1A
 
 | |
6P1C
 
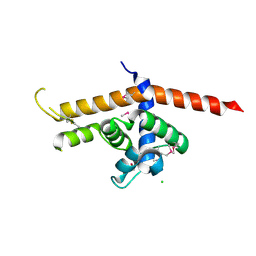 | |
5VPC
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-II crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M.C, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPB
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-I crystal | | Descriptor: | CHLORIDE ION, Protein fosB, Transcription factor jun-D | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
5VPD
 
 | | Transcription factor FosB/JunD bZIP domain in its oxidized form, type-III crystal | | Descriptor: | CHLORIDE ION, Protein fosB, SODIUM ION, ... | | Authors: | Yin, Z, Machius, M, Rudenko, G. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Activator Protein-1: redox switch controlling structure and DNA-binding.
Nucleic Acids Res., 45, 2017
|
|
8CA5
 
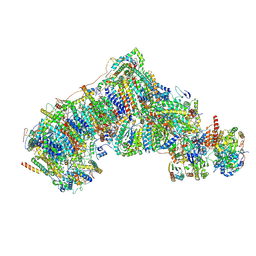 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 3). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA4
 
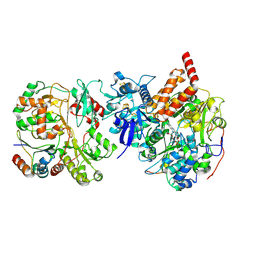 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2 N-domain). | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8C2S
 
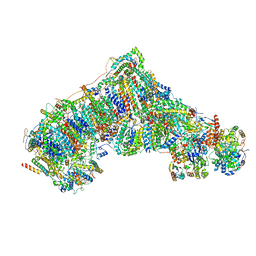 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 1). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
8CA3
 
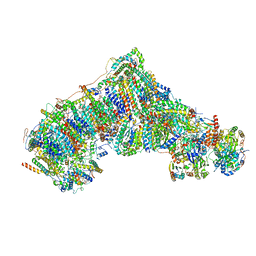 | | Cryo-EM structure NDUFS4 knockout complex I from Mus musculus heart (Class 2). | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Yin, Z, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-01-24 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into respiratory complex I deficiency and assembly from the mitochondrial disease-related ndufs4 -/- mouse.
Embo J., 43, 2024
|
|
