9KY5
 
 | | Structure of y+LAT2 bound with Arg | | Descriptor: | ARGININE, Amino acid transporter heavy chain SLC3A2, Y+L amino acid transporter 2 | | Authors: | Yan, R.H, Dai, L, Zhang, T. | | Deposit date: | 2024-12-08 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of y+LAT2 bound with Arg
To Be Published
|
|
9LDR
 
 | |
8J8L
 
 | | Overall structure of the LAT1-4F2hc bound with L-dopa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-DIHYDROXYPHENYLALANINE, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-17 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with L-dopa
To Be Published
|
|
8IDA
 
 | | Overall structure of the LAT1-4F2hc bound with tyrosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-02-12 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tyrosine
To Be Published
|
|
8J8M
 
 | | Overall structure of the LAT1-4F2hc bound with tryptophan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, Large neutral amino acids transporter small subunit 1, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-05-02 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with tryptophan
To Be Published
|
|
8JZB
 
 | | Langya Henipavirus attachment protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Yan, R.H, Guo, Y.Y. | | Deposit date: | 2023-07-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A special homotetrameric architecture of langya virus glycoprotein
To Be Published
|
|
8X0W
 
 | | Overall structure of the LAT1-4F2hc bound with Leu | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amino acid transporter heavy chain SLC3A2, LEUCINE, ... | | Authors: | Yan, R.H, Li, Y.N, Shi, T.H. | | Deposit date: | 2023-11-06 | | Release date: | 2024-11-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Overall structure of the LAT1-4F2hc bound with L-dopa
To Be Published
|
|
8WK6
 
 | | Human AGT1-rBAT complex in the apo-state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amino acid transporter heavy chain SLC3A1, CALCIUM ION, ... | | Authors: | Yan, R.H, Shi, T.H, Dong, J. | | Deposit date: | 2023-09-27 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Human AGT1-rBAT complex in the apo-state
To Be Published
|
|
8WVZ
 
 | |
8WWA
 
 | |
8WW6
 
 | |
8WW8
 
 | |
8WW9
 
 | |
8WW7
 
 | |
8XA4
 
 | | A35 of MPXV in complex with mAb981 | | Descriptor: | Heavy chain of mAb981, Light chain of mAb981, Protein OPG161 | | Authors: | Li, Y.N, Shi, T.H, Yan, R.H. | | Deposit date: | 2023-12-01 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of MPXV A35 with a neutralizing and protective human monoclonal antibody
To Be Published
|
|
7LO1
 
 | |
6NET
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus substrate complex | | Descriptor: | 2,4-dihydroxy-3,6-dimethylbenzaldehyde, CHLORIDE ION, FAD-dependent monooxygenase tropB, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NEU
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus R206Q variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NEV
 
 | | FAD-dependent monooxygenase TropB from T. stipitatus Y239F Variant | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6NES
 
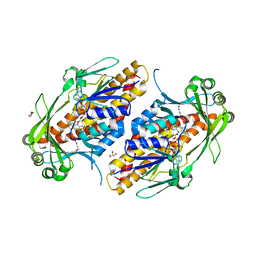 | | FAD-dependent monooxygenase TropB from T. stipitatus | | Descriptor: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Brooks, C.L, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | Deposit date: | 2018-12-18 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
6U1R
 
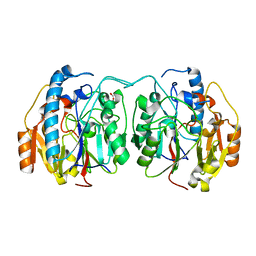 | | SxtG an amidinotransferase from the Microseira wollei in Saxitoxin biosynthetic pathway | | Descriptor: | FORMIC ACID, SxtG | | Authors: | Mallik, L, Lukowski, A.L, Narayan, A.R.H, Koutmos, M. | | Deposit date: | 2019-08-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Substrate Promiscuity of a Paralytic Shellfish Toxin Amidinotransferase.
Acs Chem.Biol., 15, 2020
|
|
7C2L
 
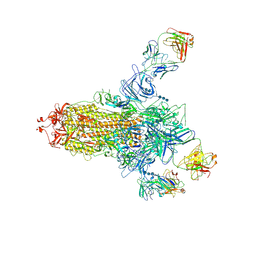 | | S protein of SARS-CoV-2 in complex bound with 4A8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Guo, Y.Y, Li, Y.N, Xia, L, Zhou, Q. | | Deposit date: | 2020-05-08 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2.
Science, 369, 2020
|
|
6DS5
 
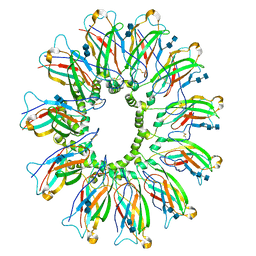 | | Cryo EM structure of human SEIPIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Seipin | | Authors: | Yan, R.H, Qian, H.W, Yan, N, Yang, H.Y. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-24 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Human SEIPIN Binds Anionic Phospholipids.
Dev. Cell, 47, 2018
|
|
6IRS
 
 | | human LAT1-4F2hc complex incubated with JPH203 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4F2 cell-surface antigen heavy chain, ... | | Authors: | Yan, R.H, Zhao, X, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature, 568, 2019
|
|
6IRT
 
 | | human LAT1-4F2hc complex bound with BCH | | Descriptor: | (1S,2S,4R)-2-aminobicyclo[2.2.1]heptane-2-carboxylic acid, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhao, X, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human LAT1-4F2hc heteromeric amino acid transporter complex.
Nature, 568, 2019
|
|
