2MGZ
 
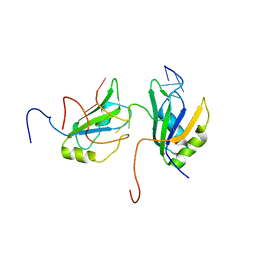 | | Solution structure of RBFOX family ASD-1 RRM and SUP-12 RRM in ternary complex with RNA | | Descriptor: | Protein ASD-1, isoform a, Protein SUP-12, ... | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2EX6
 
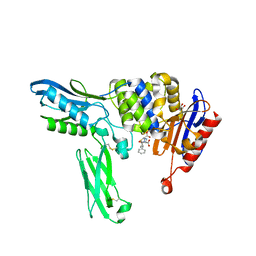 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with ampicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
1IV4
 
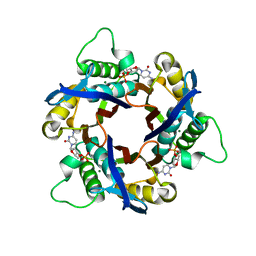 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form Substrate) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV3
 
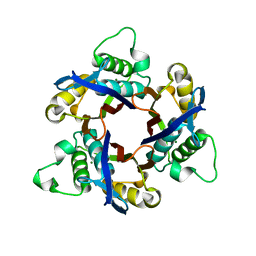 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form MG atoms) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV2
 
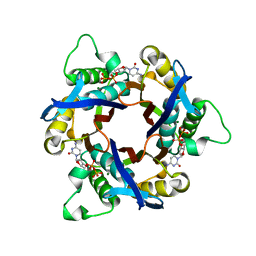 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase (bound form CDP) | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IV1
 
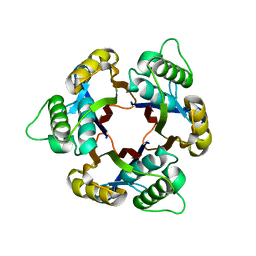 | | Structure of 2C-Methyl-D-erythritol-2,4-cyclodiphosphate Synthase | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | | Authors: | Kishida, H, Wada, T, Unzai, S, Kuzuyama, T, Terada, T, Sirouzu, M, Yokoyama, S, Tame, J.R.H, Park, S.-Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-11 | | Release date: | 2002-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2RU3
 
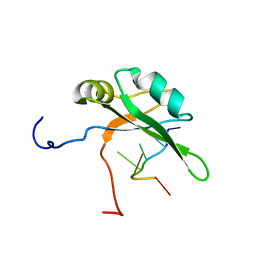 | | Solution structure of c.elegans SUP-12 RRM in complex with RNA | | Descriptor: | Protein SUP-12, isoform a, RNA (5'-R(*GP*UP*GP*UP*GP*C)-3') | | Authors: | Takahashi, M, Kuwasako, K, Unzai, S, Tsuda, K, Yoshikawa, S, He, F, Kobayashi, N, Guntert, P, Shirouzu, M, Ito, T, Tanaka, A, Yokoyama, S, Hagiwara, M, Kuroyanagi, H, Muto, Y. | | Deposit date: | 2013-11-12 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | RBFOX and SUP-12 sandwich a G base to cooperatively regulate tissue-specific splicing
Nat.Struct.Mol.Biol., 21, 2014
|
|
2RRU
 
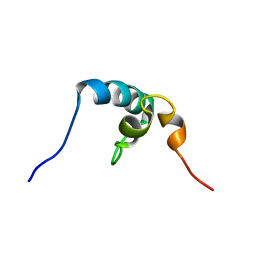 | | Solution structure of the UBA omain of p62 and its interaction with ubiquitin | | Descriptor: | Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal structure of the UBA omain of p62 and its interaction with ubiquitin
To be Published
|
|
2RPB
 
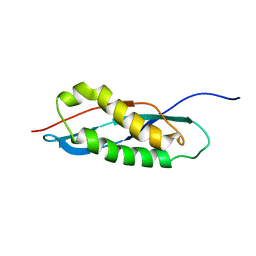 | |
2DGE
 
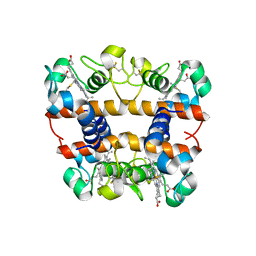 | | Crystal structure of oxidized cytochrome C6A from Arabidopsis thaliana | | Descriptor: | Cytochrome c6, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | Authors: | Chida, H, Yokoyama, T, Kawai, F, Nakazawa, A, Akazaki, H, Takayama, Y, Hirano, T, Suruga, K, Satoh, T, Yamada, S, Kawachi, R, Unzai, S, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2006-03-11 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of oxidized cytochrome c(6A) from Arabidopsis thaliana
Febs Lett., 580, 2006
|
|
1UIU
 
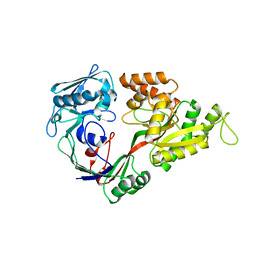 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel unliganded form) | | Descriptor: | Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIV
 
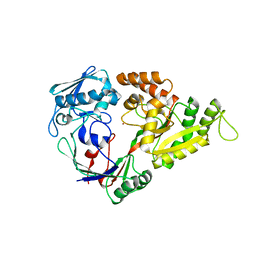 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel liganded form) | | Descriptor: | NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1V4W
 
 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH7.5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V9F
 
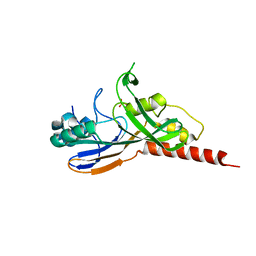 | | Crystal structure of catalytic domain of pseudouridine synthase RluD from Escherichia coli | | Descriptor: | PHOSPHATE ION, Ribosomal large subunit pseudouridine synthase D | | Authors: | Mizutani, K, Machida, Y, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
1V4U
 
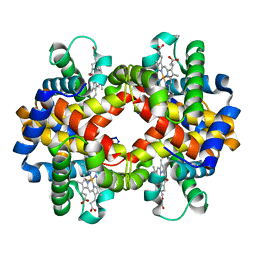 | | Crystal structure of bluefin tuna carbonmonoxy-hemoglobin | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, ... | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V4X
 
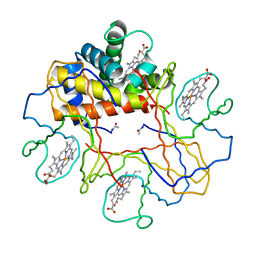 | | Crystal structure of bluefin tuna hemoglobin deoxy form at pH5.0 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, hemoglobin alpha chain, hemoglobin beta chain | | Authors: | Yokoyama, T, Chong, K.T, Miyazaki, Y, Nakatsukasa, T, Unzai, S, Miyazaki, G, Morimoto, H, Jeremy, R.H.T, Park, S.Y. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel Mechanisms of pH Sensitivity in Tuna Hemoglobin: A STRUCTURAL EXPLANATION OF THE ROOT EFFECT
J.Biol.Chem., 279, 2004
|
|
1V9K
 
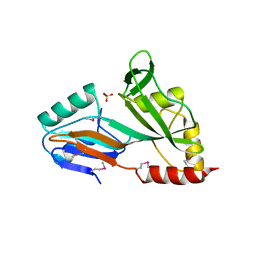 | | The crystal structure of the catalytic domain of pseudouridine synthase RluC from Escherichia coli | | Descriptor: | Ribosomal large subunit pseudouridine synthase C, SULFATE ION | | Authors: | Machida, Y, Mizutani, K, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domains of pseudouridine synthases RluC and RluD from Escherichia coli
Biochemistry, 43, 2004
|
|
3ABE
 
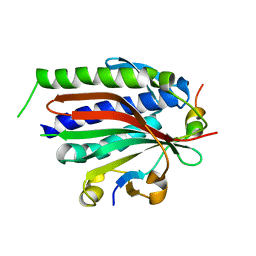 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
3ABD
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a monoclinic crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
2EB4
 
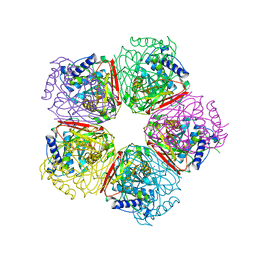 | | Crystal structure of apo-HpcG | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, SODIUM ION, THIOCYANATE ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EB5
 
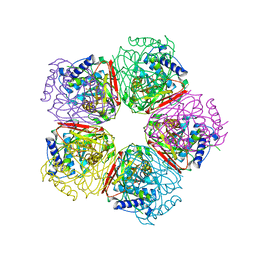 | | Crystal structure of HpcG complexed with oxalate | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EB6
 
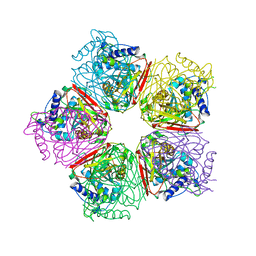 | | Crystal structure of HpcG complexed with Mg ion | | Descriptor: | 2-oxo-hept-3-ene-1,7-dioate hydratase, MAGNESIUM ION | | Authors: | Izumi, A, Rea, D, Adachi, T, Unzai, S, Park, S.Y, Roper, D.I, Tame, J.R.H. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure and Mechanism of HpcG, a Hydratase in the Homoprotocatechuate Degradation Pathway of Escherichia coli
J.Mol.Biol., 370, 2007
|
|
2EX2
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli | | Descriptor: | GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EXA
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with FAROM | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
2EX8
 
 | | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, complexed with penicillin-G | | Descriptor: | OPEN FORM - PENICILLIN G, Penicillin-binding protein 4 | | Authors: | Kishida, H, Unzai, S, Roper, D.I, Lloyd, A, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of penicillin binding protein 4 (dacB) from Escherichia coli, both in the native form and covalently linked to various antibiotics
Biochemistry, 45, 2006
|
|
