4P76
 
 | | Cellular response to a crystal-forming protein | | Descriptor: | Photoconvertible fluorescent protein, SODIUM ION | | Authors: | Tsutsui, H, Jinno, Y, Shoda, K, Tomita, A, Matsuda, M, Yamashita, E, Katayama, H, Nakagawa, A, Miyawaki, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A diffraction-quality protein crystal processed as an autophagic cargo
Mol.Cell, 58, 2015
|
|
1XSS
 
 | | Semi-rational engineering of a green-emitting coral fluorescent protein into an efficient highlighter. | | Descriptor: | MAGNESIUM ION, SODIUM ION, fluorescent protein | | Authors: | Tsutsui, H, Karasawa, S, Shimizu, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2004-10-20 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Semi-rational engineering of a coral fluorescent protein into an efficient highlighter
Embo Rep., 6, 2005
|
|
2DDD
 
 | | Unique behavior of a histidine responsible for an engineered green-to-red photoconversion process | | Descriptor: | MAGNESIUM ION, SODIUM ION, photoconvertible fluorescent protein | | Authors: | Shimizu, H, Tsutsui, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The E1 mechanism in photo-induced beta-elimination reactions for green-to-red conversion of fluorescent proteins
Chem.Biol., 16, 2009
|
|
2DDC
 
 | | Unique behavior of a histidine responsible for an engineered green-to-red photoconversion process | | Descriptor: | MAGNESIUM ION, SODIUM ION, photoconvertible fluorescent protein | | Authors: | Shimizu, H, Tsutsui, H, Nukina, N, Miyawaki, A. | | Deposit date: | 2006-01-27 | | Release date: | 2006-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The E1 mechanism in photo-induced beta-elimination reactions for green-to-red conversion of fluorescent proteins.
Chem.Biol., 16, 2009
|
|
3AW0
 
 | | Structure of SARS 3CL protease with peptidic aldehyde inhibitor | | Descriptor: | 3C-Like Proteinase, peptide ACE-SER-ALA-VAL-LEU-HIS-H | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3AW1
 
 | | Structure of SARS 3CL protease auto-proteolysis resistant mutant in the absent of inhibitor | | Descriptor: | 3C-Like Proteinase | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3ATW
 
 | | Structure-Based Design, Synthesis, Evaluation of Peptide-mimetic SARS 3CL Protease Inhibitors | | Descriptor: | 3C-Like Proteinase, peptide ACE-THR-VAL-ALC-HIS-H | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-01-20 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
3AVZ
 
 | | Structure of SARS 3CL protease with peptidic aldehyde inhibitor containing cyclohexyl side chain | | Descriptor: | 3C-Like Proteinase, peptide ACE-SER-ALA-VAL-ALC-HIS-H | | Authors: | Akaji, K, Konno, H, Mitsui, H, Teruya, K, Hattori, Y, Ozaki, T, Kusunoki, M, Sanjho, A. | | Deposit date: | 2011-03-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
1IU4
 
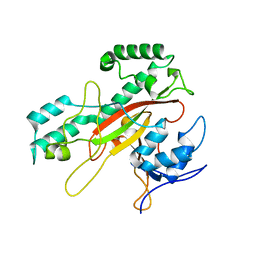 | | Crystal Structure Analysis of the Microbial Transglutaminase | | Descriptor: | microbial transglutaminase | | Authors: | Kashiwagi, T, Yokoyama, K, Ishikawa, K, Ono, K, Ejima, D, Matsui, H, Suzuki, E. | | Deposit date: | 2002-02-27 | | Release date: | 2002-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of microbial transglutaminase from Streptoverticillium mobaraense
J.Biol.Chem., 277, 2002
|
|
6DCG
 
 | | Discovery of MK-8353: An Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology | | Descriptor: | (3S)-3-(methylsulfanyl)-1-(2-{4-[4-(1-methyl-1H-1,2,4-triazol-3-yl)phenyl]-3,6-dihydropyridin-1(2H)-yl}-2-oxoethyl)-N-(3-{6-[(propan-2-yl)oxy]pyridin-3-yl}-1H-indazol-5-yl)pyrrolidine-3-carboxamide, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Boga, S.B, Deng, Y, Zhu, L, Nan, Y, Cooper, A, Shipps Jr, G.W, Doll, R, Shih, N, Zhu, H, Sun, R, Wang, T, Paliwal, S, Tsui, H, Gao, X, Yao, X, Desai, J, Wang, J, Alhassan, A.B, Kelly, J, Patel, M, Muppalla, K, Gudipati, S, Zhang, L, Buevich, A, Hesk, D, Carr, D, Dayananth, P, Mei, H, Cox, K, Sherborne, B, Hruza, A.W, Xiao, L, Jin, W, Long, B, Liu, G, Taylor, S.A, Kirschmeier, P, Windsor, W.T, Bishop, R, Samatar, A.A. | | Deposit date: | 2018-05-06 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | MK-8353: Discovery of an Orally Bioavailable Dual Mechanism ERK Inhibitor for Oncology.
ACS Med Chem Lett, 9, 2018
|
|
2EK9
 
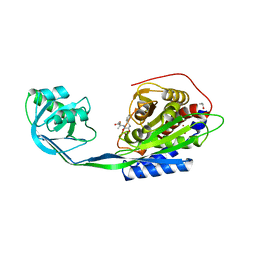 | |
2EK8
 
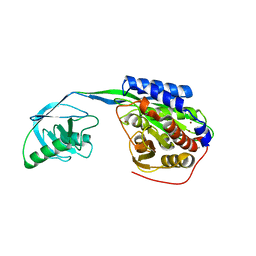 | |
6LVY
 
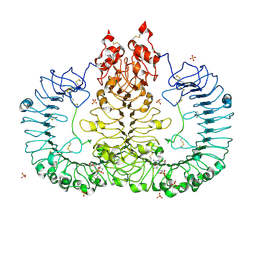 | | Crystal structure of TLR7/Cpd-2 (SM-360320) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LW0
 
 | | Crystal structure of TLR7/Cpd-6 (DSR-139293) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purin-6-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LVZ
 
 | | Crystal structure of TLR7/Cpd-3 (SM-394830) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(pyridin-3-ylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LVX
 
 | | Crystal structure of TLR7/Cpd-1 (SM-374527) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-butoxy-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LW1
 
 | | Cryo-EM structure of TLR7/Cpd-7 (DSR-139970) complex in open form | | Descriptor: | 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-6-methyl-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purine, Toll-like receptor 7 | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
4PF3
 
 | | Mineralocorticoid receptor ligand-binding domain with compuond 37a | | Descriptor: | 1,2-ETHANEDIOL, 6-[1-(2,2-difluoro-3-hydroxypropyl)-5-(4-fluorophenyl)-3-methyl-1H-pyrazol-4-yl]-2H-1,4-benzoxazin-3(4H)-one, Mineralocorticoid receptor | | Authors: | Sogabe, S, Habuka, N. | | Deposit date: | 2014-04-28 | | Release date: | 2014-11-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of 6-[5-(4-fluorophenyl)-3-methyl-pyrazol-4-yl]-benzoxazin-3-one derivatives as novel selective nonsteroidal mineralocorticoid receptor antagonists
Bioorg.Med.Chem., 22, 2014
|
|
1G0D
 
 | | CRYSTAL STRUCTURE OF RED SEA BREAM TRANSGLUTAMINASE | | Descriptor: | PROTEIN-GLUTAMINE GAMMA-GLUTAMYLTRANSFERASE, SULFATE ION | | Authors: | Noguchi, K, Ishikawa, K, Yokoyama, K, Ohtsuka, T, Nio, N, Suzuki, E. | | Deposit date: | 2000-10-06 | | Release date: | 2001-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of red sea bream transglutaminase.
J.Biol.Chem., 276, 2001
|
|
8JL8
 
 | | Crystal structure of the collagen binding domain of Cnm from Streptococcus mutans | | Descriptor: | Collagen-binding adhesin, GLYCEROL, SULFATE ION | | Authors: | Tanaka, S.-i, Hirata, A, Takano, K. | | Deposit date: | 2023-06-02 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure, Stability and Binding Properties of Collagen-Binding Domains from Streptococcus mutans.
Chemistry, 5, 2023
|
|
3VW5
 
 | |
3WKH
 
 | | Crystal structure of cellobiose 2-epimerase in complex with epilactose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WY4
 
 | | Crystal structure of alpha-glucosidase mutant E271Q in complex with maltose | | Descriptor: | Alpha-glucosidase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
3WKG
 
 | | Crystal structure of cellobiose 2-epimerase in complex with glucosylmannose | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
3WKI
 
 | | Crystal structure of cellobiose 2-epimerase in complex with cellobiitol | | Descriptor: | CHLORIDE ION, Cellobiose 2-epimerase, PHOSPHATE ION, ... | | Authors: | Fujiwara, T, Saburi, W, Tanaka, I, Yao, M. | | Deposit date: | 2013-10-21 | | Release date: | 2013-12-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Structural Insights into the Epimerization of beta-1,4-Linked Oligosaccharides Catalyzed by Cellobiose 2-Epimerase, the Sole Enzyme Epimerizing Non-anomeric Hydroxyl Groups of Unmodified Sugars
J.Biol.Chem., 289, 2014
|
|
