6ZCT
 
 | | Nonstructural protein 10 (nsp10) from SARS CoV-2 | | Descriptor: | ZINC ION, nsp10 | | Authors: | Rogstam, A, Nyblom, M, Christensen, S, Sele, C, Lindvall, T, Rasmussen, A.A, Andre, I, Fisher, S.Z, Knecht, W, Kozielski, F. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Non-Structural Protein 10 from Severe Acute Respiratory Syndrome Coronavirus-2.
Int J Mol Sci, 21, 2020
|
|
2XF4
 
 | | Crystal structure of Salmonella enterica serovar typhimurium YcbL | | Descriptor: | HYDROXYACYLGLUTATHIONE HYDROLASE, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Stamp, A, Owen, P, El Omari, K, Nichols, C, Lockyer, M, Lamb, H, Charles, I, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Salmonella Enterica Serovar Typhimurium Ycbl: An Unusual Type II Glyoxalase
Protein Sci., 19, 2010
|
|
3AWI
 
 | | Bifunctional tRNA modification enzyme MnmC from Escherichia coli | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, tRNA 5-methylaminomethyl-2-thiouridine biosynthesis bifunctional protein mnmC | | Authors: | Kitamura, A, Sengoku, T, Nishimoto, M, Yokoyama, S, Bessho, Y. | | Deposit date: | 2011-03-23 | | Release date: | 2011-06-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the bifunctional tRNA modification enzyme MnmC from Escherichia coli
Protein Sci., 2011
|
|
2DX3
 
 | |
2DX2
 
 | |
2DX4
 
 | |
1F7T
 
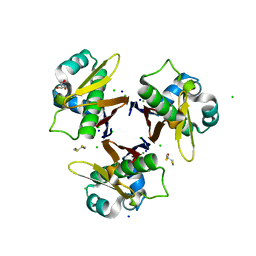 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE AT 1.8A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1F7L
 
 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH COENZYME A AT 1.5A | | Descriptor: | CALCIUM ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-27 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1F80
 
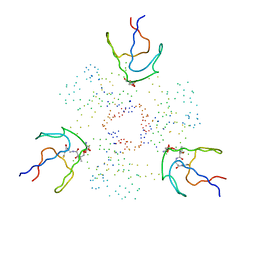 | | HOLO-(ACYL CARRIER PROTEIN) SYNTHASE IN COMPLEX WITH HOLO-(ACYL CARRIER PROTEIN) | | Descriptor: | ACYL CARRIER PROTEIN, HOLO-(ACYL CARRIER PROTEIN) SYNTHASE, SODIUM ION | | Authors: | Parris, K.D, Lin, L, Tam, A, Mathew, R, Hixon, J, Stahl, M, Fritz, C.C, Seehra, J, Somers, W.S. | | Deposit date: | 2000-06-28 | | Release date: | 2001-06-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of substrate binding to Bacillus subtilis holo-(acyl carrier protein) synthase reveal a novel trimeric arrangement of molecules resulting in three active sites.
Structure Fold.Des., 8, 2000
|
|
1HY8
 
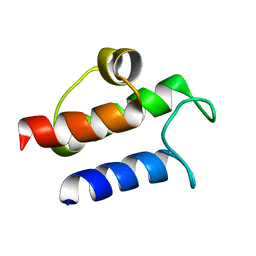 | | SOLUTION STRUCTURE OF B. SUBTILIS ACYL CARRIER PROTEIN | | Descriptor: | ACYL CARRIER PROTEIN | | Authors: | Xu, G.-Y, Tam, A, Lin, L, Hixon, J, Fritz, C.C, Power, R. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of B. subtilis acyl carrier protein.
Structure, 9, 2001
|
|
3VSS
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain complexed with fructose | | Descriptor: | Beta-fructofuranosidase, beta-D-fructofuranose | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
3VSR
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase catalytic domain | | Descriptor: | Beta-fructofuranosidase | | Authors: | Tonozuka, T, Tamaki, A, Yokoi, G, Miyazaki, T, Ichikawa, M, Nishikawa, A, Ohta, Y, Hidaka, Y, Katayama, K, Hatada, Y, Ito, T, Fujita, K. | | Deposit date: | 2012-05-08 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a lactosucrose-producing enzyme, Arthrobacter sp. K-1 beta-fructofuranosidase
Enzyme.Microb.Technol., 51, 2012
|
|
8C87
 
 | | Double mutant A(L172)C/L(L246)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Yukhimchuk, D. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C7C
 
 | | Double mutant V(M84)C/A(L278)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-14 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C6K
 
 | | Double mutant A(L53)C/I(L64)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-12 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C88
 
 | | Double mutant G(M19)C/T(L214)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Yukhimchuk, D. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
8C5X
 
 | | Double mutant A(L37)C/S(L99)C structure of Photosynthetic Reaction Center From Cereibacter sphaeroides strain RV | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, BACTERIOCHLOROPHYLL A, ... | | Authors: | Gabdulkhakov, A, Selikhanov, G, Fufina, T, Vasilieva, L, Atamas, A, Uhimchuk, D. | | Deposit date: | 2023-01-10 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Stabilization of Cereibacter sphaeroides Photosynthetic Reaction Center by the Introduction of Disulfide Bonds.
Membranes (Basel), 13, 2023
|
|
4J12
 
 | | monomeric Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, human Fc fragment | | Authors: | Ishino, T, Wang, M, Mosyak, L, Tam, A, Duan, W, Svenson, K, Joyce, A, O'Hara, D, Lin, L, Somers, W, Kriz, R. | | Deposit date: | 2013-01-31 | | Release date: | 2013-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering a Monomeric Fc Domain Modality by N-Glycosylation for the Half-life Extension of Biotherapeutics.
J.Biol.Chem., 288, 2013
|
|
2HNY
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HNZ
 
 | | Crystal Structure of E138K Mutant HIV-1 Reverse Transcriptase in Complex with PETT-2 | | Descriptor: | 1-[2-(4-ETHOXY-3-FLUOROPYRIDIN-2-YL)ETHYL]-3-(5-METHYLPYRIDIN-2-YL)THIOUREA, PHOSPHATE ION, Reverse transcriptase/ribonuclease H | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-13 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2HND
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Stamp, A, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into mechanisms of non-nucleoside drug resistance for HIV-1 reverse transcriptases mutated at codons 101 or 138.
Febs J., 273, 2006
|
|
2KFQ
 
 | | NMR Structure of FP1 | | Descriptor: | FP1 | | Authors: | Araki, M, Tamura, A. | | Deposit date: | 2009-02-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solubility-dependent structural formation of a 25-residue, natively unfolded protein, induced by addition of a seven-residue peptide fragment
Febs J., 276, 2009
|
|
2LCF
 
 | | Solution structure of GppNHp-bound H-RasT35S mutant protein | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Araki, M, Shima, F, Yoshikawa, Y, Muraoka, S, Ijiri, Y, Nagahara, Y, Shirono, T, Kataoka, T, Tamura, A. | | Deposit date: | 2011-04-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers.
J.Biol.Chem., 286, 2011
|
|
5I4N
 
 | |
4YUR
 
 | | Crystal Structure of Plk4 Kinase Domain Bound to Centrinone | | Descriptor: | 2-({2-fluoro-4-[(2-fluoro-3-nitrobenzyl)sulfonyl]phenyl}sulfanyl)-5-methoxy-N-(3-methyl-1H-pyrazol-5-yl)-6-(morpholin-4-yl)pyrimidin-4-amine, Serine/threonine-protein kinase PLK4 | | Authors: | Shiau, A.K, Motamedi, A. | | Deposit date: | 2015-03-19 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4.
Science, 348, 2015
|
|
