2R2I
 
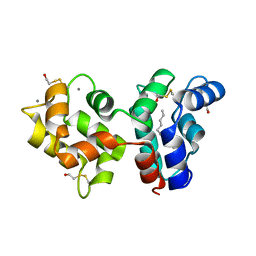 | | Myristoylated Guanylate Cyclase Activating Protein-1 with Calcium Bound | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, Guanylyl cyclase-activating protein 1, ... | | Authors: | Stephen, R. | | Deposit date: | 2007-08-25 | | Release date: | 2007-12-11 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stabilizing function for myristoyl group revealed by the crystal structure of a neuronal calcium sensor, guanylate cyclase-activating protein 1.
Structure, 15, 2007
|
|
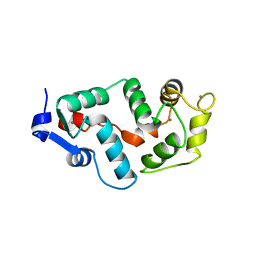
2GGZ
 
 | |
1X9A
 
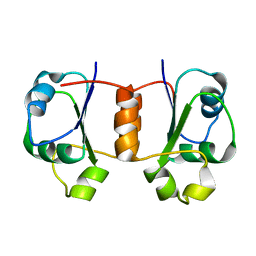 | | Solution NMR Structure of Protein Tm0979 from Thermotoga maritima. Ontario Center for Structural Proteomics Target TM0979_1_87; Northeast Structural Genomics Consortium Target VT98. | | Descriptor: | hypothetical protein TM0979 | | Authors: | Gaspar, J.A, Liu, C, Vassall, K.A, Stathopulos, P.B, Meglei, G, Stephen, R, Pineda-Lucena, A, Wu, B, Yee, A, Arrowsmith, C.H, Meiering, E.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-08-20 | | Release date: | 2004-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel member of the YchN-like fold: solution structure of the hypothetical protein Tm0979 from Thermotoga maritima.
Protein Sci., 14, 2005
|
|
2F9B
 
 | | Discovery of Novel Heterocyclic Factor VIIa Inhibitors | | Descriptor: | Coagulation factor VII, Tissue factor, {5-(5-AMINO-1H-PYRROLO[3,2-B]PYRIDIN-2-YL)-6-HYDROXY-3'-NITRO-BIPHENYL-3-YL]-ACETIC ACID | | Authors: | Rai, R, Kolesnikov, A, Sprengeler, P.A, Torkelson, S, Ton, T, Katz, B.A, Yu, C, Hendrix, J, Shrader, W.D, Stephens, R, Cabuslay, R, Sanford, E, Young, W.B. | | Deposit date: | 2005-12-05 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Discovery of novel heterocyclic factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5G1V
 
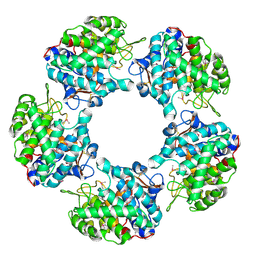 | | Linalool Dehydratase Isomerase: Selenomethionine Derivative | | Descriptor: | LINALOOL DEHYDRATASE ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2017-02-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1W
 
 | | Apo Structure of Linalool Dehydratase-Isomerase | | Descriptor: | 1,2-ETHANEDIOL, LINALOOL DEHYDRATASE/ISOMERASE, METHYLMALONIC ACID | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
5G1U
 
 | | Linalool Dehydratase Isomerase in complex with Geraniol | | Descriptor: | Geraniol, LINALOOL DEHYDRATASE/ISOMERASE | | Authors: | Chambers, S, Hau, A, Man, H, Omar, M, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2016-03-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural and functional insights into asymmetric enzymatic dehydration of alkenols.
Nat. Chem. Biol., 13, 2017
|
|
6WGH
 
 | |
6PQ3
 
 | | Crystal structure of GDP-bound KRAS with ten residues long internal tandem duplication in the switch II region | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Dharmaiah, S, Chan, A.H, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2019-07-08 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | RASinternal tandem duplication disrupts GTPase-activating protein (GAP) binding to activate oncogenic signaling.
J.Biol.Chem., 295, 2020
|
|
3G86
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with thiazine inhibitor | | Descriptor: | N-{3-[6-fluoro-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, NICKEL (II) ION, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones
Bioorg.Med.Chem.Lett., 19, 2009
|
|
