5LAE
 
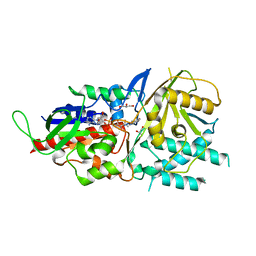 | | Crystal structure of murine N1-acetylpolyamine oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Snijder, A, Barlind, L. | | Deposit date: | 2016-06-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
1HCM
 
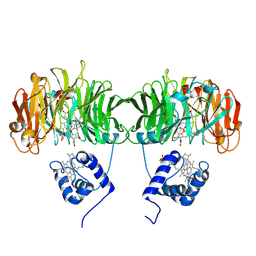 | |
5LFO
 
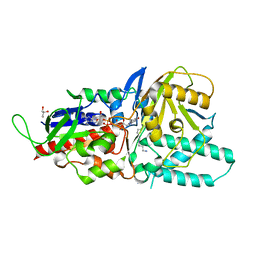 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-04 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5LGB
 
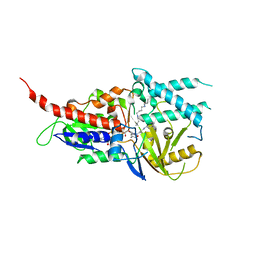 | | Crystal structure of murine N1-acetylpolyamine oxidase in complex with MDL72527 | | Descriptor: | FAD-MDL72527 adduct, N,N'-BIS(2,3-BUTADIENYL)-1,4-BUTANE-DIAMINE, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase,Peroxisomal N(1)-acetyl-spermine/spermidine oxidase | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-07-06 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
5MBX
 
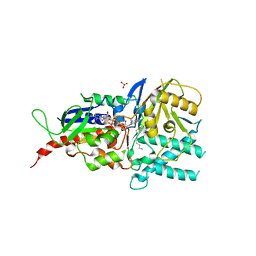 | | Crystal structure of reduced murine N1-acetylpolyamine oxidase in complex with N1-acetylspermine | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N-[3-({4-[(3-aminopropyl)amino]butyl}amino)propyl]acetamide, Peroxisomal N(1)-acetyl-spermine/spermidine oxidase, ... | | Authors: | Sjogren, T, Aagaard, A, Wassvik, C, Snijder, A, Barlind, L. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Structure of Murine N(1)-Acetylspermine Oxidase Reveals Molecular Details of Vertebrate Polyamine Catabolism.
Biochemistry, 56, 2017
|
|
1DY7
 
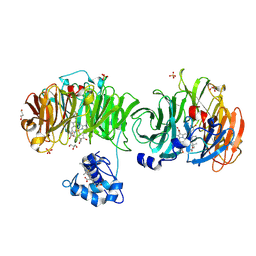 | | Cytochrome cd1 Nitrite Reductase, CO complex | | Descriptor: | CARBON MONOXIDE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Svensson-Ek, M, Hajdu, J, Brzezinski, P. | | Deposit date: | 2000-01-28 | | Release date: | 2000-09-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proton-Coupled Structural Changes Upon Binding of Carbon Monoxide to Cytochrome Cd(1): A Combined Flash Photolysis and X-Ray Crystallography Study
Biochemistry, 39, 2000
|
|
1H9Y
 
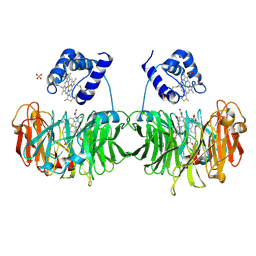 | |
1H9X
 
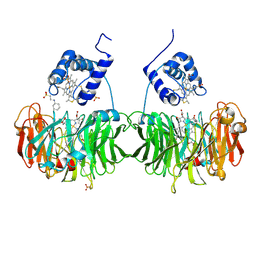 | | Cytochrome cd1 Nitrite Reductase, reduced form | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYTOCHROME CD1 NITRITE REDUCTASE, HEME C, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-08-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Alternative Form of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase
J.Biol.Chem., 276, 2001
|
|
1HJ4
 
 | |
1HJ3
 
 | | Cytochrome cd1 Nitrite Reductase, dioxygen complex | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
1HJ5
 
 | | Cytochrome cd1 Nitrite Reductase, reoxidised enzyme | | Descriptor: | GLYCEROL, HEME C, HEME D, ... | | Authors: | Sjogren, T, Hajdu, J. | | Deposit date: | 2001-01-08 | | Release date: | 2001-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structure of the bound dioxygen species in the cytochrome oxidase reaction of cytochrome cd1 nitrite reductase.
J. Biol. Chem., 276, 2001
|
|
4AL1
 
 | | Crystal structure of Human PS-1 GSH-analog complex | | Descriptor: | L-gamma-glutamyl-S-(2-biphenyl-4-yl-2-oxoethyl)-L-cysteinylglycine, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AL0
 
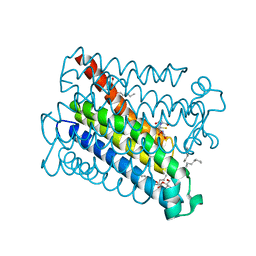 | | Crystal structure of Human PS-1 | | Descriptor: | GLUTATHIONE, PALMITIC ACID, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Sjogren, T, Nord, J, Ek, M, Johansson, P, Liu, G, Geschwindner, S. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal Structure of Microsomal Prostaglandin E2 Synthase Provides Insight Into Diversity in the Mapeg Superfamily.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1GQ1
 
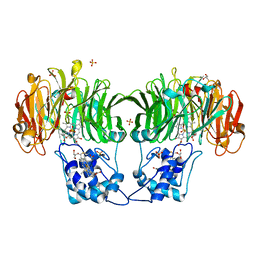 | | CYTOCHROME CD1 NITRITE REDUCTASE, Y25S mutant, OXIDISED FORM | | Descriptor: | CYTOCHROME CD1 NITRITE REDUCTASE, GLYCEROL, HEME C, ... | | Authors: | Sjogren, T, Gordon, E.H.J, Lofqvist, M, Richter, C.D, Hajdu, J, Ferguson, S.J. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Kinetic Properties of Paracoccus Pantotrophus Cytochrome Cd1 Nitrite Reductase with the D1 Heme Active Site Ligand Tyrosine 25 Replaced by Serine
J.Biol.Chem., 278, 2003
|
|
7NI3
 
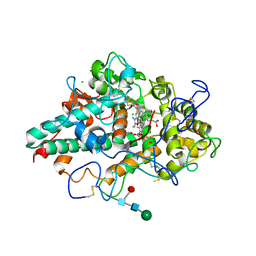 | | CRYSTAL STRUCTURE OF NATIVE HUMAN MYELOPEROXIDASE IN COMPLEX WITH CPD 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-sulfanylidene-3-[(2R)-tetrahydro-2-furanylmethyl]-1,2,3,7-tetrahydro-6H-purin-6-one, CALCIUM ION, ... | | Authors: | Sjogren, T, Inghardt, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of AZD4831, a Mechanism-Based Irreversible Inhibitor of Myeloperoxidase, As a Potential Treatment for Heart Failure with Preserved Ejection Fraction.
J.Med.Chem., 65, 2022
|
|
7NI1
 
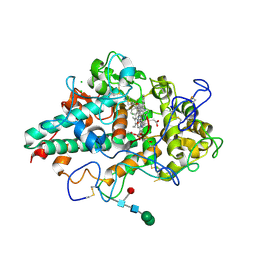 | | CRYSTAL STRUCTURE OF NATIVE HUMAN MYELOPEROXIDASE IN COMPLEX WITH CPD 9 | | Descriptor: | (S)-1-(2-(amino(phenyl)methyl)benzyl)-2-thioxo-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sjogren, T, Inghardt, T. | | Deposit date: | 2021-02-11 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of AZD4831, a Mechanism-Based Irreversible Inhibitor of Myeloperoxidase, As a Potential Treatment for Heart Failure with Preserved Ejection Fraction.
J.Med.Chem., 65, 2022
|
|
2V0M
 
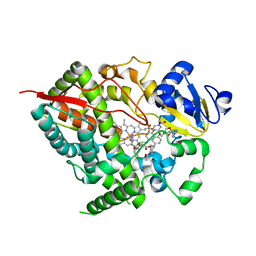 | | Crystal structure of human P450 3A4 in complex with ketoconazole | | Descriptor: | 1-ACETYL-4-(4-{[(2S,4R)-2-(2,4-DICHLOROPHENYL)-2-(1H-IMIDAZOL-1-YLMETHYL)-1,3-DIOXOLAN-4-YL]METHOXY}PHENYL)PIPERAZINE, CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sjogren, T, Ekroos, M. | | Deposit date: | 2007-05-15 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Ligand Promiscuity in Cytochrome P450 3A4
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2J0D
 
 | |
4UV4
 
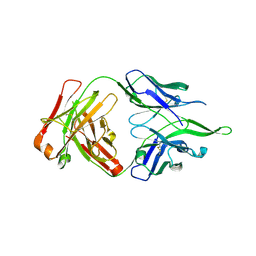 | | Crystal structure of anti-FPR Fpro0165 Fab fragment | | Descriptor: | FPRO0165 FAB | | Authors: | Douthwaite, J.A, Sridharan, S, Huntington, C, Marwood, R, Hammersley, J, Hakulinen, J.K, Ek, M, Sjogren, T, Rider, D, Privezentzev, C, Seaman, J.C, Cariuk, P, Knights, V, Young, J, Wilkinson, T, Sleeman, M, Finch, D.K, Lowe, D.C, Vaughan, T.J. | | Deposit date: | 2014-08-04 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Affinity Maturation of a Novel Antagonistic Human Monoclonal Antibody with a Long Vh Cdr3 Targeting the Class a Gpcr Formyl-Peptide Receptor 1.
Mabs, 7, 2015
|
|
3ZS0
 
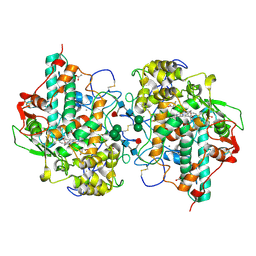 | | Human Myeloperoxidase inactivated by TX2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-FLUOROBENZYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
5ALB
 
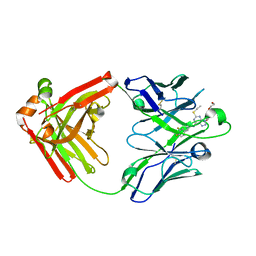 | | Ticagrelor antidote candidate MEDI2452 in complex with ticagrelor | | Descriptor: | MEDI2452 HEAVY CHAIN, MEDI2452 LIGHT CHAIN, Ticagrelor | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
5ALC
 
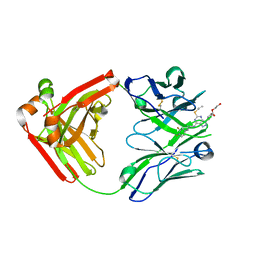 | | Ticagrelor antidote candidate Fab 72 in complex with ticagrelor | | Descriptor: | ANTI-TICAGRELOR FAB 72, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Buchanan, A, Newton, P, Pehrsson, S, Inghardt, T, Antonsson, T, Svensson, P, Sjogren, T, Oster, L, Janefeldt, A, Sandinge, A, Keyes, F, Austin, M, Spooner, J, Penney, M, Howells, G, Vaughan, T, Nylander, S. | | Deposit date: | 2015-03-07 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterisation of a Specific Antidote for Ticagrelor.
Blood, 125, 2015
|
|
3ZS1
 
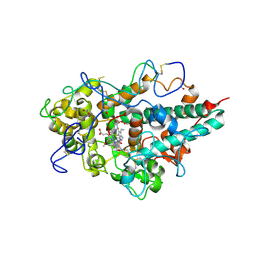 | | Human Myeloperoxidase inactivated by TX5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-METHOXYETHYL)-2-THIOXO-1,2,3,7-TETRAHYDRO-6H-PURIN-6-ONE, ACETATE ION, ... | | Authors: | Tiden, A.K, Sjogren, T, Svensson, M, Bernlind, A, Senthilmohan, R, Auchere, F, Norman, H, Markgren, P.O, Gustavsson, S, Schmidt, S, Lundquist, S, Forbes, L.V, Magon, N.J, Jameson, G.N, Eriksson, H, Kettle, A.J. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Thioxanthines are Mechanism-Based Inactivators of Myeloperoxidase that Block Oxidative Stress During Inflammation.
J.Biol.Chem., 286, 2011
|
|
4NY4
 
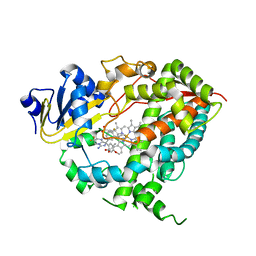 | | Crystal structure of CYP3A4 in complex with an inhibitor | | Descriptor: | (8R)-3,3-difluoro-8-[4-fluoro-3-(pyridin-3-yl)phenyl]-8-(4-methoxy-3-methylphenyl)-2,3,4,8-tetrahydroimidazo[1,5-a]pyrimidin-6-amine, Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Branden, G, Sjogren, T, Xue, Y. | | Deposit date: | 2013-12-10 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure-based ligand design to overcome CYP inhibition in drug discovery projects.
Drug Discov Today, 19, 2014
|
|
4NZ2
 
 | | Crystal structure of CYP2C9 in complex with an inhibitor | | Descriptor: | (2R)-N-{4-[(3-bromophenyl)sulfonyl]-2-chlorophenyl}-3,3,3-trifluoro-2-hydroxy-2-methylpropanamide, Cytochrome P450 2C9, GLYCEROL, ... | | Authors: | Branden, G, Sjogren, T, Xue, Y. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-based ligand design to overcome CYP inhibition in drug discovery projects.
Drug Discov Today, 19, 2014
|
|
