5JZB
 
 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZ9
 
 | | Crystal structure of HsaD bound to 3,5-dichloro-4-hydroxybenzenesulphonic acid | | Descriptor: | 3,5-dichloro-4-hydroxybenzene-1-sulfonic acid, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZS
 
 | | HsaD bound to 3,5-dichloro-4-hydroxybenzoic acid | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase BphD, 3,5-dichloro-4-hydroxybenzoic acid | | Authors: | Ryan, A, Polycarpou, E, Lack, N, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E.D, Ballet, R, Abuhammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-17 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
3R6W
 
 | | paAzoR1 binding to nitrofurazone | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Ryan, A, Kaplan, K, Laurieri, N, Lowe, E, Sim, E. | | Deposit date: | 2011-03-22 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Activation of nitrofurazone by azoreductases: multiple activities in one enzyme.
Sci Rep, 1, 2011
|
|
4B55
 
 | | Crystal Structure of the Covalent Adduct Formed between Mycobacterium marinum Aryalamine N-acetyltransferase and Phenyl vinyl ketone a derivative of Piperidinols | | Descriptor: | 3-hydroxy-1-phenylpropan-1-one, ARYLAMINE N-ACETYLTRANSFERASE NAT | | Authors: | Abuhammad, A, Fullam, E, Lowe, E.D, Staunton, D, Kawamura, A, Westwood, I.M, Bhakta, S, Garner, A.C, Wilson, D.L, Seden, P.T, Davies, S.G, Russell, A.J, Garman, E.F, Sim, E. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Piperidinols that Show Anti-Tubercular Activity as Inhibitors of Arylamine N-Acetyltransferase: An Essential Enzyme for Mycobacterial Survival Inside Macrophages.
Plos One, 7, 2012
|
|
4N65
 
 | | Crystal structure of paAzoR1 bound to anthraquinone-2-sulphonate | | Descriptor: | 9,10-dioxo-9,10-dihydroanthracene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Kaplan, E, Crescente, V, Lowe, E, Preston, G.M, Sim, E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Identification of NAD(P)H Quinone Oxidoreductase Activity in Azoreductases from P. aeruginosa: Azoreductases and NAD(P)H Quinone Oxidoreductases Belong to the Same FMN-Dependent Superfamily of Enzymes.
Plos One, 9, 2014
|
|
4N9Q
 
 | | Crystal structure of paAzoR1 bound to ubiquinone-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, GLYCEROL, ... | | Authors: | Ryan, A, Kaplan, E, Crescente, V, Lowe, E, Preston, G.M, Sim, E. | | Deposit date: | 2013-10-21 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of NAD(P)H Quinone Oxidoreductase Activity in Azoreductases from P. aeruginosa: Azoreductases and NAD(P)H Quinone Oxidoreductases Belong to the Same FMN-Dependent Superfamily of Enzymes.
Plos One, 9, 2014
|
|
3LT5
 
 | | X-ray Crystallographic structure of a Pseudomonas Aeruginosa Azoreductase in complex with balsalazide | | Descriptor: | (3E)-3-({4-[(2-carboxyethyl)carbamoyl]phenyl}hydrazono)-6-oxocyclohexa-1,4-diene-1-carboxylic acid, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Ryan, A, Laurieri, N, Westwood, I, Wang, C.-J, Lowe, E, Sim, E. | | Deposit date: | 2010-02-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Novel Mechanism for Azoreduction
J.Mol.Biol., 400, 2010
|
|
3LTW
 
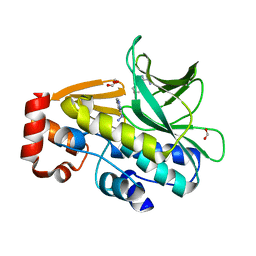 | | The structure of mycobacterium marinum arylamine n-acetyltransferase in complex with hydralazine | | Descriptor: | 1-hydrazinophthalazine, Arylamine N-acetyltransferase Nat, FORMIC ACID | | Authors: | Abuhammad, A.M, Lowe, E.D, Fullam, E, Noble, M, Garman, E.F, Sim, E. | | Deposit date: | 2010-02-16 | | Release date: | 2010-07-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the architecture of the Mycobacterium marinum arylamine N-acetyltransferase active site
Protein Cell, 1, 2010
|
|
3KEG
 
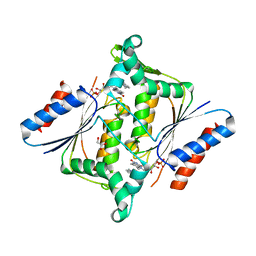 | | X-ray Crystallographic Structure of a Y131F mutant of Pseudomonas Aeruginosa Azoreductase in complex with Methyl RED | | Descriptor: | 2-(4-DIMETHYLAMINOPHENYL)DIAZENYLBENZOIC ACID, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase 1, ... | | Authors: | Wang, C.-J, Laurieri, N, Abuhammad, A, Lowe, E, Westwood, I, Ryan, A, Sim, E. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of tyrosine 131 in the active site of paAzoR1, an azoreductase with specificity for the inflammatory bowel disease prodrug balsalazide
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4BGF
 
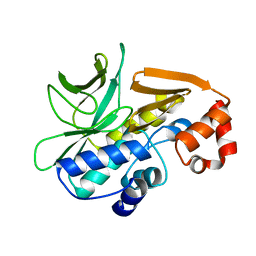 | | The 3D-structure of arylamine-N-acetyltransferase from M. tuberculosis | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE NAT | | Authors: | Abuhammad, A, Lowe, E.D, McDonough, M.A, Shaw Stewart, P.D, Kolek, S.A, Sim, E, Garman, E.F. | | Deposit date: | 2013-03-26 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure of arylamine N-acetyltransferase from Mycobacterium tuberculosis determined by cross-seeding with the homologous protein from M. marinum: triumph over adversity.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
1E2T
 
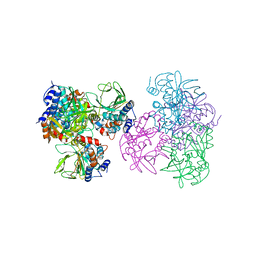 | | Arylamine N-acetyltransferase (NAT) from Salmonella typhimurium | | Descriptor: | N-HYDROXYARYLAMINE O-ACETYLTRANSFERASE | | Authors: | Sinclair, J.C, Sandy, J, Delgoda, R, Sim, E, Noble, M.E.M. | | Deposit date: | 2000-05-24 | | Release date: | 2000-07-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Arylamine N-Acetyltransferase Reveals a Catalytic Triad
Nat.Struct.Biol., 7, 2000
|
|
2WUG
 
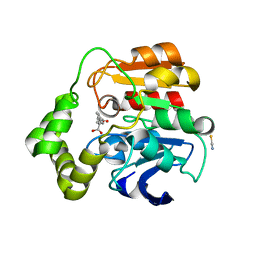 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R.L, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a C-C Hydrolase from Mycobacterium Tuberuclosis Involved in Cholesterol Metabolism.
J.Biol.Chem., 285, 2010
|
|
2WUE
 
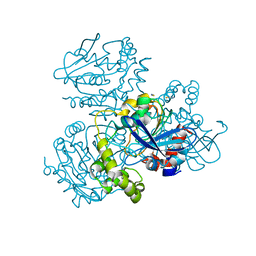 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with HOPODA | | Descriptor: | (3E,5R)-8-(2-CHLOROPHENYL)-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, THIOCYANATE ION | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
2WUF
 
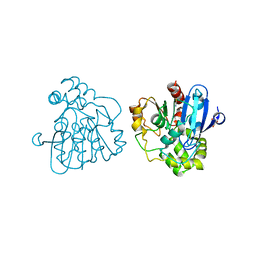 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis in complex with 4,9DSHA | | Descriptor: | (3E,5R)-8-[(1S,3AR,4R,7AS)-1-HYDROXY-7A-METHYL-5-OXOOCTAHYDRO-1H-INDEN-4-YL]-5-METHYL-2,6-DIOXOOCT-3-ENOATE, 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, ... | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
2WUD
 
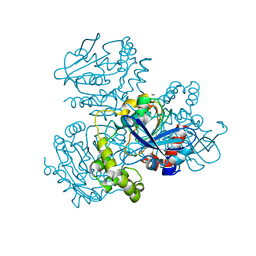 | | Crystal structure of S114A mutant of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, THIOCYANATE ION | | Authors: | Lack, N.A, Yam, K.C, Lowe, E.D, Horsman, G.P, Owen, R, Sim, E, Eltis, L.D. | | Deposit date: | 2009-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism.
J. Biol. Chem., 285, 2010
|
|
2XW7
 
 | | Structure of Mycobacterium smegmatis putative reductase MS0308 | | Descriptor: | DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, ... | | Authors: | Evangelopoulos, D, Gupta, A, Lack, N, Cronin, N, Daviter, T, Sim, E, Keep, N.H, Bhakta, S. | | Deposit date: | 2010-11-01 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of an Oxidoreductase from the Arylamine N-Acetyltransferase Operon in Mycobacterium Smegmatis.
FEBS J., 278, 2011
|
|
1W6F
 
 | | Arylamine N-acetyltransferase from Mycobacterium smegmatis with the anti-tubercular drug isoniazid bound in the active site. | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Holton, S, Fullham, E, Sim, E, Noble, M.E.M. | | Deposit date: | 2004-08-17 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Anti-Tubercular Drug Isoniazid to the Arylamine N-Acetyltransferase Protein from Mycobacterium Smegmatis
Protein Sci., 14, 2005
|
|
1W5R
 
 | | X-ray crystallographic structure of a C70Q Mycobacterium smegmatis N- arylamine Acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Holton, S.J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E.M, Sim, E. | | Deposit date: | 2004-08-09 | | Release date: | 2005-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Investigation of the Catalytic Triad of Arylamine N-Acetyltransferases: Essential Residues Required for Acetyl Transfer to Arylamines.
Biochem.J., 390, 2005
|
|
2BSZ
 
 | | Structure of Mesorhizobium loti arylamine N-acetyltransferase 1 | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE 1 | | Authors: | Holton, S.J, Dairou, J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Noble, M.E.M, Sim, E. | | Deposit date: | 2005-05-24 | | Release date: | 2005-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Mesorhizobium Loti Arylamine N-Acetyltransferase 1.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1W4T
 
 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
1GX3
 
 | | M. smegmatis arylamine N-acetyl transferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Mushtaq, A, Kawamura, A, Sinclair, J, Sim, E, Noble, M. | | Deposit date: | 2002-03-26 | | Release date: | 2002-06-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of Arylamine N-Acetyltransferase from Mycobacterium Smegmatis-an Enzyme which Inactivates the Anti-Tubercular Drug, Isoniazid
J.Mol.Biol., 318, 2002
|
|
2VFC
 
 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase in complex with CoA | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE, COENZYME A | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
2VFB
 
 | | The structure of Mycobacterium marinum arylamine N-acetyltransferase | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Fullam, E, Westwood, I.M, Anderton, M.C, Lowe, E.D, Sim, E, Noble, M.E.M. | | Deposit date: | 2007-11-02 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Divergence of Cofactor Recognition Across Evolution: Coenzyme a Binding in a Prokaryotic Arylamine N-Acetyltransferase.
J.Mol.Biol., 375, 2008
|
|
2VF2
 
 | | X-ray crystal structure of HsaD from Mycobacterium tuberculosis | | Descriptor: | 2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOATE HYDROLASE BPHD, GLYCEROL, SULFATE ION | | Authors: | Lack, N, Lowe, E.D, Liu, J, Eltis, L.D, Noble, M.E.M, Sim, E, Westwood, I.M. | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Hsad, a Steroid-Degrading Hydrolase, from Mycobacterium Tuberculosis.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
