6YHF
 
 | | Solution NMR Structure of APP TMD | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-29 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHO
 
 | | Solution NMR Structure of APP G38P mutant TM | | Descriptor: | Amyloid-beta precursor protein G38P mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHP
 
 | | Solution NMR Structure of APP V44M mutant TMD | | Descriptor: | Amyloid-beta precursor protein V44M mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHX
 
 | | Solution NMR Structure of APP I45T mutant TMD | | Descriptor: | Amyloid-beta precursor protein I45T mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
6YHI
 
 | | Solution NMR Structure of APP G38L mutant TMD | | Descriptor: | Amyloid-beta precursor protein G38L mutant | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2020-03-30 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Altered Hinge Conformations in APP Transmembrane Helix Mutants May Affect Enzyme-Substrate Interactions of gamma-Secretase.
Acs Chem Neurosci, 11, 2020
|
|
3FCD
 
 | |
7YYI
 
 | |
7QAP
 
 | | Three-dimensional structure of the PGAM5 G17L mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7QAO
 
 | | Three-dimensional structure of the PGAM5 C12S mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7QAL
 
 | | Three-dimensional structure of the PGAM5 C12L mutant TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7QAM
 
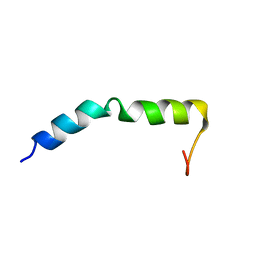 | | Three-dimensional structure of the PGAM5 WT TMD | | Descriptor: | Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Silber, M, Muhle-Goll, C. | | Deposit date: | 2021-11-17 | | Release date: | 2022-05-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state.
J.Biol.Chem., 298, 2022
|
|
7Z0B
 
 | |
7Z07
 
 | |
7Z08
 
 | |
3L3S
 
 | |
8RPQ
 
 | |
8RQ6
 
 | |
6G2S
 
 | | Crystal structure of FimH in complex with a pentaflourinated biphenyl alpha D-mannoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-(hydroxymethyl)-6-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]phenoxy]oxane-3,4,5-triol, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
6G2R
 
 | | Crystal structure of FimH in complex with a tetraflourinated biphenyl alpha D-mannoside | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]-2,3,5,6-tetrakis(fluoranyl)benzenecarbonitrile, SULFATE ION, ... | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
5MCA
 
 | | Crystal structure of FimH-LD R60P variant in the apo state | | Descriptor: | Protein FimH, SULFATE ION | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Conformational switch of the bacterial adhesin FimH in the absence of the regulatory domain: Engineering a minimalistic allosteric system.
J. Biol. Chem., 293, 2018
|
|
5MUC
 
 | | Crystal structure of the FimH lectin domain in complex with 1,5-Anhydromannitol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, Protein FimH | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | KinITC-One Method Supports both Thermodynamic and Kinetic SARs as Exemplified on FimH Antagonists.
Chemistry, 24, 2018
|
|
5L4W
 
 | | Crystal structure of FimH lectin domain in complex with 3-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 3-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4X
 
 | | Crystal structure of FimH lectin domain in complex with 4-Deoxy-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-deoxy-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4U
 
 | | Crystal structure of FimH lectin domain in complex with 2-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 2-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
5L4Y
 
 | | Crystal structure of FimH lectin domain in complex with 4-Fluoro-Heptylmannoside | | Descriptor: | Protein FimH, heptyl 4-deoxy-4-fluoro-alpha-D-mannopyranoside | | Authors: | Jakob, R.P, Zihlmann, P, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2016-05-26 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-Affinity Carbohydrate-Lectin Interactions: How Nature Makes it Possible
To Be Published
|
|
