1D9N
 
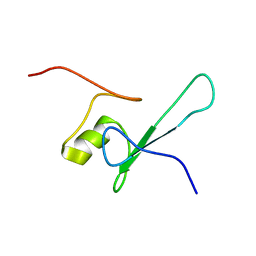 | | SOLUTION STRUCTURE OF THE METHYL-CPG-BINDING DOMAIN OF THE METHYLATION-DEPENDENT TRANSCRIPTIONAL REPRESSOR MBD1/PCM1 | | Descriptor: | METHYL-CPG-BINDING PROTEIN MBD1 | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Nakao, M, Shirakawa, M. | | Deposit date: | 1999-10-28 | | Release date: | 2000-10-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1.
EMBO J., 18, 1999
|
|
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
1KLV
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor associated protein | | Descriptor: | GABA(A) Receptor associated protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-13 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
3ASK
 
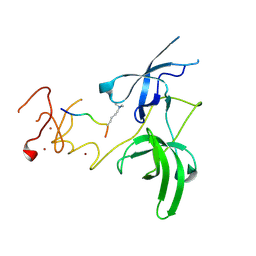 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ZINC ION | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2013-06-05 | | Method: | X-RAY DIFFRACTION (2.904 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1OCP
 
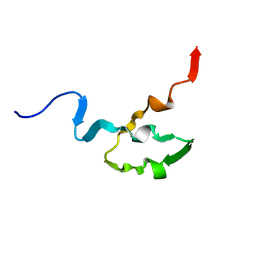 | | SOLUTION STRUCTURE OF OCT3 POU-HOMEODOMAIN | | Descriptor: | OCT-3 | | Authors: | Morita, E.H, Hayashi, F, Shirakawa, M, Kyogoku, Y. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Oct-3 POU-Homeodomain in Solution, as Determined by Triple Resonance Heteronuclear Multidimensional NMR Spectroscopy
Protein Sci., 4, 1995
|
|
1UGL
 
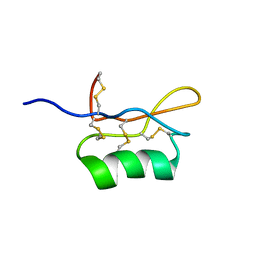 | | Solution structure of S8-SP11 | | Descriptor: | S-locus pollen protein | | Authors: | Mishima, M, Takayama, S, Sasaki, K, Jee, J.G, Kojima, C, Isogai, A, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Male Determinant Factor for Brassica Self-incompatibility
J.Biol.Chem., 278, 2003
|
|
3A1A
 
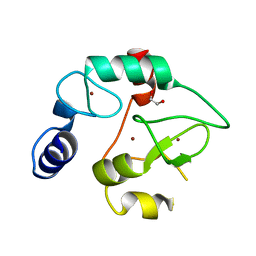 | | Crystal Structure of the DNMT3A ADD domain | | Descriptor: | 1,2-ETHANEDIOL, DNA (cytosine-5)-methyltransferase 3A, ZINC ION | | Authors: | Otani, J, Arita, K, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2009-03-28 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain
Embo Rep., 10, 2009
|
|
2D2P
 
 | | The solution structure of micelle-bound peptide | | Descriptor: | Pituitary adenylate cyclase activating polypeptide-38 | | Authors: | Tateishi, Y, Jee, J.G, Inooka, H, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The solution structure of micelle-bound peptide
To be Published
|
|
1UEL
 
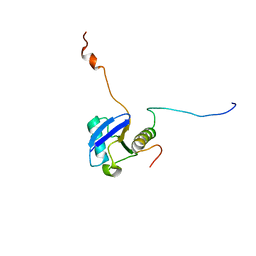 | | Solution structure of ubiquitin-like domain of hHR23B complexed with ubiquitin-interacting motif of proteasome subunit S5a | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, UV excision repair protein RAD23 homolog B | | Authors: | Fujiwara, K, Tenno, T, Jee, J.G, Sugasawa, K, Ohki, I, Kojima, C, Tochio, H, Hiroaki, H, Hanaoka, H, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-19 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-interacting Motif of S5a Bound to the Ubiquitin-like Domain of HR23B
J.Biol.Chem., 279, 2004
|
|
3ASL
 
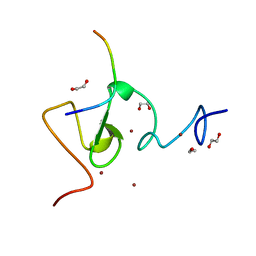 | | Structure of UHRF1 in complex with histone tail | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1, Histone H3.3, ... | | Authors: | Arita, K, Sugita, K, Unoki, M, Hamamoto, R, Sekiyama, N, Tochio, H, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2010-12-16 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8HFP
 
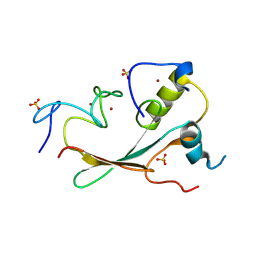 | | Crystal structure of the methyl-CpG-binding domain of SETDB2 in complex with the cysteine-rich domain of C11orf46 protein | | Descriptor: | ARL14 effector protein, Histone-lysine N-methyltransferase SETDB2, SULFATE ION, ... | | Authors: | Mahana, Y, Ariyoshi, M, Shirakawa, M. | | Deposit date: | 2022-11-11 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural evidence for protein-protein interaction between the non-canonical methyl-CpG-binding domain of SETDB proteins and C11orf46.
Structure, 32, 2024
|
|
2D07
 
 | | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase | | Descriptor: | G/T mismatch-specific thymine DNA glycosylase, Ubiquitin-like protein SMT3B | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-07-26 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of SUMO-3-modified Thymine-DNA Glycosylase
J.Mol.Biol., 359, 2006
|
|
1KM7
 
 | | Solution Structure and Backbone Dynamics of GABARAP, GABAA Receptor Associated Protein | | Descriptor: | GABA(A) Receptor Associated Protein | | Authors: | Kouno, T, Miura, K, Tada, M, Kanematsu, T, Tate, S, Shirakawa, M, Hirata, M, Kawano, K. | | Deposit date: | 2001-12-14 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and '5N resonance assignments of GABARAP, GABAA receptor associated protein.
J.Biomol.Nmr, 22, 2002
|
|
3B0F
 
 | | Crystal structure of the UBA domain of p62 and its interaction with ubiquitin | | Descriptor: | SULFATE ION, Sequestosome-1 | | Authors: | Isogai, S, Morimoto, D, Arita, K, Unzai, S, Tenno, T, Hasegawa, J, Sou, Y, Komatsu, M, Tanaka, K, Shirakawa, M, Tochio, H. | | Deposit date: | 2011-06-09 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the ubiquitin-associated (UBA) domain of p62 and its interaction with ubiquitin.
J.Biol.Chem., 286, 2011
|
|
1A2B
 
 | | HUMAN RHOA COMPLEXED WITH GTP ANALOGUE | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA | | Authors: | Ihara, K, Muraguchi, S, Kato, M, Shimizu, T, Shirakawa, M, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1997-12-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue.
J.Biol.Chem., 273, 1998
|
|
1ED7
 
 | | SOLUTION STRUCTURE OF THE CHITIN-BINDING DOMAIN OF BACILLUS CIRCULANS WL-12 CHITINASE A1 | | Descriptor: | CHITINASE A1 | | Authors: | Ikegami, T, Okada, T, Hashimoto, M, Seino, S, Watanabe, T, Shirakawa, M. | | Deposit date: | 2000-01-27 | | Release date: | 2000-05-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.
J.Biol.Chem., 275, 2000
|
|
2EI9
 
 | | Crystal structure of R1Bm endonuclease domain | | Descriptor: | ACETIC ACID, Non-LTR retrotransposon R1Bmks ORF2 protein | | Authors: | Maita, N, Aoyagi, H, Osanai, M, Shirakawa, M, Fujiwara, H. | | Deposit date: | 2007-03-12 | | Release date: | 2007-07-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the sequence specificity of the R1Bm endonuclease domain by structural and biochemical studies.
Nucleic Acids Res., 35, 2007
|
|
5ZCZ
 
 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZD0
 
 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Z4B
 
 | | GB1 structure determination in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Protein LG | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5Y3T
 
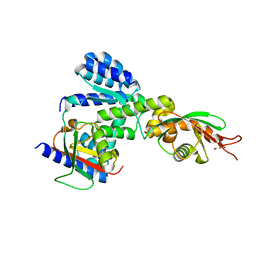 | | Crystal structure of hetero-trimeric core of LUBAC: HOIP double-UBA complexed with HOIL-1L UBL and SHARPIN UBL | | Descriptor: | E3 ubiquitin-protein ligase RNF31, GLYCEROL, RanBP-type and C3HC4-type zinc finger-containing protein 1, ... | | Authors: | Tokunaga, A, Fujita, H, Ariyoshi, M, Ohki, I, Tochio, H, Iwai, K, Shirakawa, M. | | Deposit date: | 2017-07-31 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cooperative Domain Formation by Homologous Motifs in HOIL-1L and SHARPIN Plays A Crucial Role in LUBAC Stabilization.
Cell Rep, 23, 2018
|
|
3WO4
 
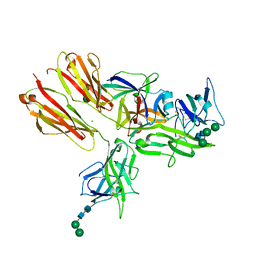 | | Crystal structure of the IL-18 signaling ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO2
 
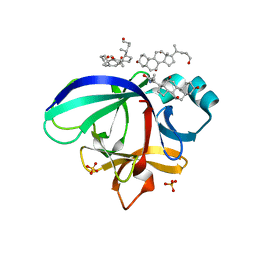 | | Crystal structure of human interleukin-18 | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Interleukin-18, SULFATE ION | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
3WO3
 
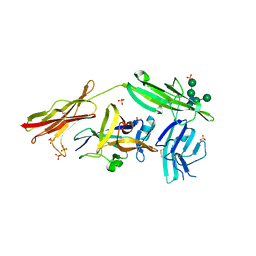 | | Crystal structure of IL-18 in complex with IL-18 receptor alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tsutsumi, N, Kimura, T, Arita, K, Ariyoshi, M, Ohnishi, H, Kondo, N, Shirakawa, M, Kato, Z, Tochio, H. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis for receptor recognition of human interleukin-18
Nat Commun, 5, 2014
|
|
1IG4
 
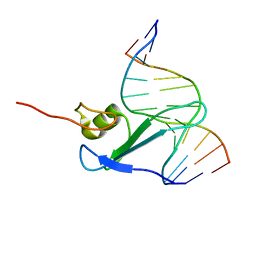 | | Solution Structure of the Methyl-CpG-Binding Domain of Human MBD1 in Complex with Methylated DNA | | Descriptor: | 5'-D(*GP*TP*AP*TP*CP*(5CM)P*GP*GP*AP*TP*AP*C)-3', Methyl-CpG Binding Protein | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Jee, J.-G, Ikegami, T, Nakao, M, Shirakawa, M. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA.
Cell(Cambridge,Mass.), 105, 2001
|
|
