1SN8
 
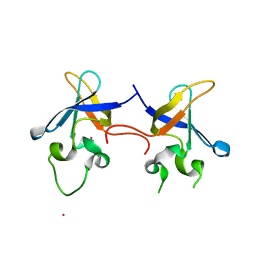 | | Crystal structure of the S1 domain of RNase E from E. coli (Pb derivative) | | Descriptor: | LEAD (II) ION, Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SMX
 
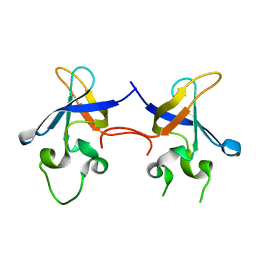 | | Crystal structure of the S1 domain of RNase E from E. coli (native) | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-09 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
1SLJ
 
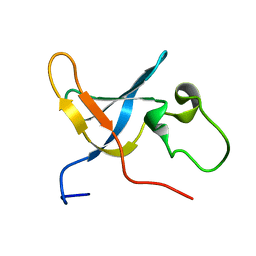 | | Solution structure of the S1 domain of RNase E from E. coli | | Descriptor: | Ribonuclease E | | Authors: | Schubert, M, Edge, R.E, Lario, P, Cook, M.A, Strynadka, N.C.J, Mackie, G.A, McIntosh, L.P. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
J.Mol.Biol., 341, 2004
|
|
2JPP
 
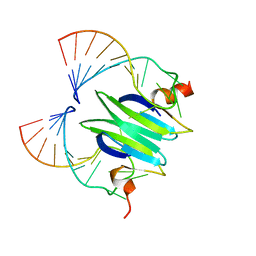 | | Structural basis of RsmA/CsrA RNA recognition: Structure of RsmE bound to the Shine-Dalgarno sequence of hcnA mRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*UP*CP*AP*CP*GP*GP*AP*UP*GP*AP*AP*GP*CP*CP*C)-3'), Translational repressor | | Authors: | Schubert, M, Lapouge, K, Duss, O, Oberstrass, F.C, Jelesarov, I, Haas, D, Allain, F.H.-T. | | Deposit date: | 2007-05-21 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Nat.Struct.Mol.Biol., 14, 2007
|
|
2LIE
 
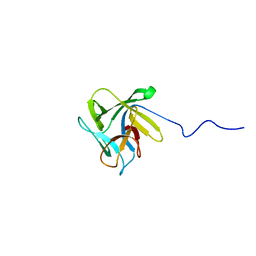 | | NMR structure of the lectin CCL2 | | Descriptor: | CCL2 lectin | | Authors: | Schubert, M, Walti, M.A, Egloff, P, Bleuler-Martinez, S, Aebi, M, Allain, F.F.-H, Kunzler, M. | | Deposit date: | 2011-08-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2LIQ
 
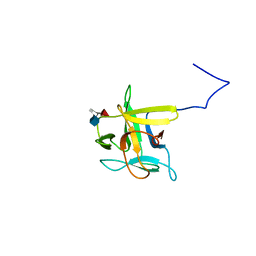 | | Solution structure of CCL2 in complex with glycan | | Descriptor: | CCL2 lectin, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]methyl 2-acetamido-2-deoxy-beta-D-glucopyranoside | | Authors: | Schubert, M, Bleuler-Martinez, S, Walti, M.A, Egloff, P, Aebi, M, Kuenzler, M, Allain, F.H.-T. | | Deposit date: | 2011-08-30 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Plasticity of the beta-Trefoil Protein Fold in the Recognition and Control of Invertebrate Predators and Parasites by a Fungal Defence System
Plos Pathog., 8, 2012
|
|
2MK1
 
 | |
4USO
 
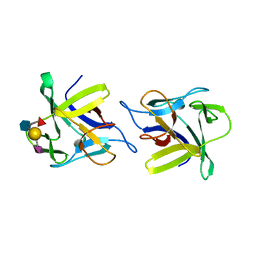 | | X-ray structure of the CCL2 lectin in complex with sialyl lewis X | | Descriptor: | CCL2 LECTIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
4USP
 
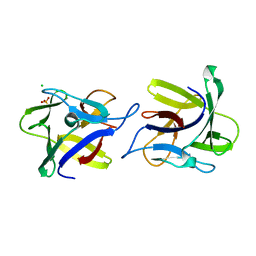 | | X-ray structure of the dimeric CCL2 lectin in native form | | Descriptor: | CCL2 LECTIN, CHLORIDE ION, PHOSPHATE ION | | Authors: | Bleuler-Martinez, S, Varrot, A, Schubert, M, Stutz, M, Sieber, R, Hengartner, M, Aebi, M, Kunzler, M. | | Deposit date: | 2014-07-11 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dimerization of the fungal defense lectin CCL2 is essential for its toxicity against nematodes.
Glycobiology, 27, 2017
|
|
8A0C
 
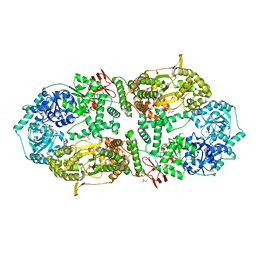 | | Capsular polysaccharide synthesis multienzyme in complex with CMP | | Descriptor: | Bcs3, CYTIDINE-5'-MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
8A0M
 
 | | Capsular polysaccharide synthesis multienzyme in complex with capsular polymer fragment | | Descriptor: | Bcs3, MAGNESIUM ION, beta-D-ribosyl-(1->1)-D-ribitol-5-phosphate | | Authors: | Cifuente, J.O, Schulze, J, Bethe, A, Di Domenico, V, Litschko, C, Budde, I, Eidenberger, L, Thiesler, H, Ramon-Roth, I, Berger, M, Claus, H, DAngelo, C, Marina, A, Gerardy-Schahn, R, Schubert, M, Guerin, M.E, Fiebig, T. | | Deposit date: | 2022-05-29 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Nat.Chem.Biol., 19, 2023
|
|
2K32
 
 | | Truncated AcrA from Campylobacter jejuni for glycosylation studies | | Descriptor: | A | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F. | | Deposit date: | 2008-04-17 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
2K33
 
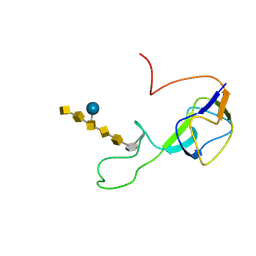 | | Solution structure of an N-glycosylated protein using in vitro glycosylation | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-[beta-D-glucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, AcrA | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F.H.-T. | | Deposit date: | 2008-04-18 | | Release date: | 2009-02-03 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
6EU9
 
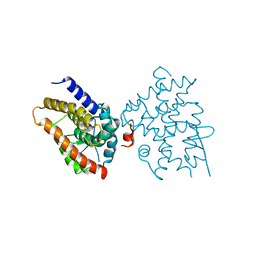 | | Crystal structure of Platynereis dumerilii RAR ligand-binding domain in complex with all-trans retinoic acid | | Descriptor: | RETINOIC ACID, Retinoic acid receptor | | Authors: | Handberg-Thorsager, M, Gutierrez-Mazariegos, J, Arold, S.T, Nadendla, E.K, Bertucci, P.Y, Germain, P, Tomancak, P, Pierzchalski, K, Jones, J.W, Albalat, R, Kane, M.A, Bourguet, W, Laudet, V, Arendt, D, Schubert, M. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation.
Sci Adv, 4, 2018
|
|
1M8M
 
 | | SOLID-STATE MAS NMR STRUCTURE OF THE A-SPECTRIN SH3 DOMAIN | | Descriptor: | SPECTRIN ALPHA CHAIN, BRAIN | | Authors: | Castellani, F, Van Rossum, B, Diehl, A, Schubert, M, Rehbein, K, Oschkinat, H. | | Deposit date: | 2002-07-25 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy
Nature, 420, 2002
|
|
2MF0
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MFF
 
 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2N7A
 
 | | Solution structure of the human Siglec-8 lectin domain | | Descriptor: | Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2MFG
 
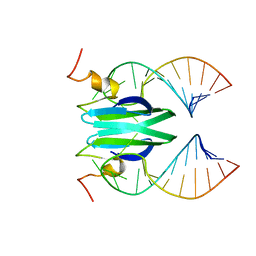 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFC
 
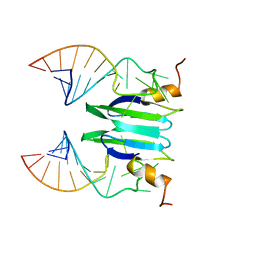 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL1)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL1(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-10 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFH
 
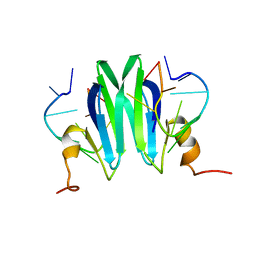 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFE
 
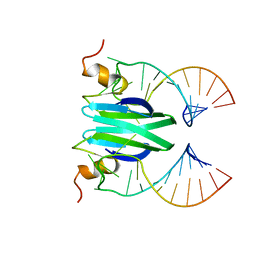 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2N7B
 
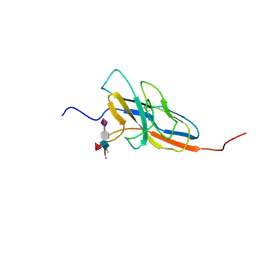 | | Solution structure of the human Siglec-8 lectin domain in complex with 6'sulfo sialyl Lewisx | | Descriptor: | 3-aminopropan-1-ol, N-acetyl-alpha-neuraminic acid-(2-3)-6-O-sulfo-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, Sialic acid-binding Ig-like lectin 8 | | Authors: | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | Deposit date: | 2015-09-07 | | Release date: | 2016-07-06 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7B3O
 
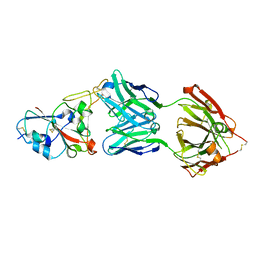 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
