1LJX
 
 | | THE STRUCTURE OF D(TPGPCPGPCPA)2 AT 293K: COMPARISON OF THE EFFECT OF SEQUENCE AND TEMPERATURE | | Descriptor: | 5'-D(*TP*GP*CP*GP*CP*A)-3', MAGNESIUM ION | | Authors: | Thiyagarajan, S, Satheesh Kumar, P, Rajan, S.S, Gautham, N. | | Deposit date: | 2002-04-23 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of d(TGCGCA)2 at 293 K: comparison of the effects of sequence and temperature.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3V7J
 
 | | Co-crystal structure of Wild Type Rat polymerase beta: Enzyme-DNA binary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*AP*AP*CP*TP*CP*AP*CP*AP*TP*A)-3'), ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3R97
 
 | | Crystal structure of malonyl-CoA:acyl carrier protein transacylase (FabD), Xoo0880, from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Malonyl CoA-ACP transacylase | | Authors: | Natarajan, S, Jung, J.W, Kang, L.W. | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of malonyl-CoA:acyl carrier protein transacylase (FabD), Xoo0880, from Xanthomonas oryzae pv. oryzae KACC10331
To be Published
|
|
3V7K
 
 | |
3V7L
 
 | | Apo Structure of Rat DNA polymerase beta K72E variant | | Descriptor: | CHLORIDE ION, DNA polymerase beta, SODIUM ION, ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
3FK5
 
 | |
3JRU
 
 | | Crystal structure of Leucyl Aminopeptidase (pepA) from Xoo0834,Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | CALCIUM ION, CARBONATE ION, Probable cytosol aminopeptidase, ... | | Authors: | Natarajan, S, Huynh, K.-H, Kang, L.W. | | Deposit date: | 2009-09-08 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Leucyl Aminopeptidase (pepA) from Xoo0834,Xanthomonas oryzae pv. oryzae KACC10331
to be published
|
|
3K89
 
 | |
3NX6
 
 | |
5D75
 
 | | Crystal structure of Human FKBD25 in complex with FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500), Peptidyl-prolyl cis-trans isomerase FKBP3 | | Authors: | Rajan, S, Prakash, A, Yoon, H.S. | | Deposit date: | 2015-08-13 | | Release date: | 2016-04-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the FK506 binding domain of human FKBP25 in complex with FK506.
Protein Sci., 25, 2016
|
|
4MGV
 
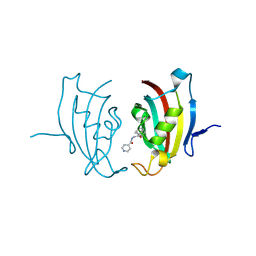 | |
4Z0X
 
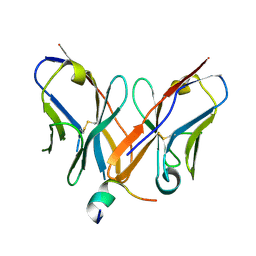 | |
4EC2
 
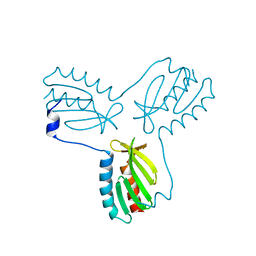 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
3OEQ
 
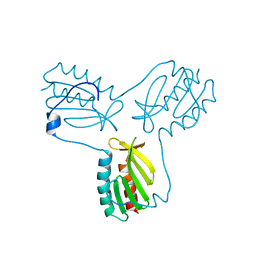 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, with full length n-terminus | | Descriptor: | Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
3OER
 
 | | Crystal structure of trimeric frataxin from the yeast saccharomyces cerevisiae, complexed with cobalt | | Descriptor: | COBALT (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Ta, C, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Oligomerization Propensity and Flexibility of Yeast Frataxin Studied by X-ray Crystallography and Small-Angle X-ray Scattering.
J.Mol.Biol., 414, 2011
|
|
6J05
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | ARSENIC, SODIUM ION, Transcriptional regulator ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
6J0E
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARSENIC, Arsenic responsive repressor ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
4E3Q
 
 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
3F72
 
 | | Crystal Structure of the Staphylococcus aureus pI258 CadC Metal Binding Site 2 Mutant | | Descriptor: | Cadmium efflux system accessory protein, SODIUM ION | | Authors: | Kandegedara, A, Thiyagarajan, S, Kondapalli, K.C, Stemmler, T.L, Rosen, B.P. | | Deposit date: | 2008-11-07 | | Release date: | 2009-04-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Role of bound Zn(II) in the CadC Cd(II)/Pb(II)/Zn(II)-responsive repressor.
J.Biol.Chem., 284, 2009
|
|
4E3R
 
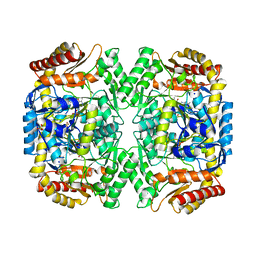 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
2HTT
 
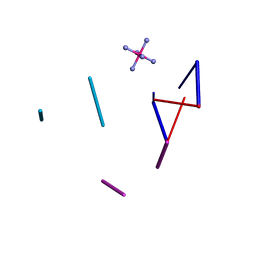 | | Ruthenium Hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DTP*DG)-3'), ... | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2HTO
 
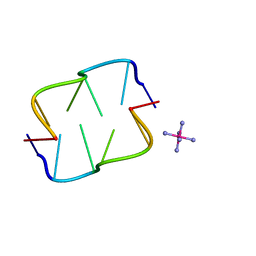 | | Ruthenium hexammine ion interactions with Z-DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DCP*DGP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DCP*DGP*DCP*DG)-3'), RUTHENIUM (III) HEXAAMINE ION | | Authors: | Bharanidharan, D, Thiyagarajan, S, Gautham, N. | | Deposit date: | 2006-07-26 | | Release date: | 2006-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Hexammineruthenium(III) ion interactions with Z-DNA
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3UQI
 
 | |
4PPI
 
 | | Crystal structure of Bcl-xL hexamer | | Descriptor: | Bcl-2-like protein 1, GLYCEROL | | Authors: | Sreekanth, R, Yoon, H.S. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
4QVF
 
 | |
