5V4K
 
 | | Crystal structure of NEDD4 LIR-fused human LC3B_2-119 | | Descriptor: | GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B,Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Qiu, Y, Zheng, Y, Schulman, B. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Insights into links between autophagy and the ubiquitin system from the structure of LC3B bound to the LIR motif from the E3 ligase NEDD4.
Protein Sci., 26, 2017
|
|
2LN0
 
 | | Structure of MOZ | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Qiu, Y. | | Deposit date: | 2011-12-15 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription.
Genes Dev., 26, 2012
|
|
2L89
 
 | |
2ARH
 
 | | Crystal Structure of a Protein of Unknown Function AQ1966 from Aquifex aeolicus VF5 | | Descriptor: | CALCIUM ION, SELENIUM ATOM, SULFATE ION, ... | | Authors: | Qiu, Y, Kim, Y, Yang, X, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-08-19 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal Structure of a Hypothetical Protein Aq_1966 from Aquifex aeolicus VF5
To be Published
|
|
1U14
 
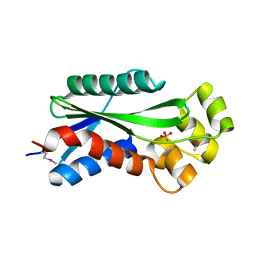 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
1R4V
 
 | | 1.9A crystal structure of protein AQ328 from Aquifex aeolicus | | Descriptor: | CACODYLATE ION, Hypothetical protein AQ_328, ZINC ION | | Authors: | Qiu, Y, Tereshko, V, Kim, Y, Zhang, R, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Proteins, 62, 2006
|
|
1S9U
 
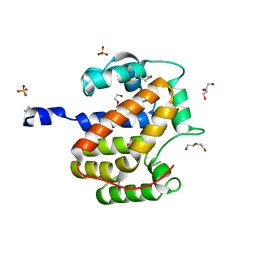 | | Atomic structure of a putative anaerobic dehydrogenase component | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, putative component of anaerobic dehydrogenases | | Authors: | Qiu, Y, Zhang, R, Tereshko, V, Kim, Y, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-05 | | Release date: | 2004-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The 1.38 A crystal structure of DmsD protein from Salmonella typhimurium, a proofreading chaperone on the Tat pathway.
Proteins, 71, 2008
|
|
1Y7R
 
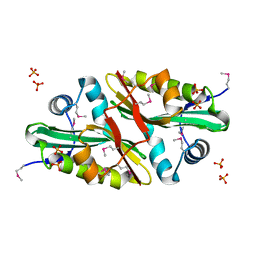 | | 1.7 A Crystal Structure of Protein of Unknown Function SA2161 from Meticillin-Resistant Staphylococcus aureus, Probable Acetyltransferase | | Descriptor: | PHOSPHATE ION, hypothetical protein SA2161 | | Authors: | Qiu, Y, Zhang, R, Collart, F, Holzle, D, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A Crystal Structure of Hypothetical Protein SA2161 from Meticillin-Resistant Staphylococcus aureus
To be Published
|
|
3V43
 
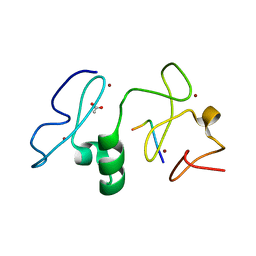 | | Crystal structure of MOZ | | Descriptor: | ACETATE ION, Histone H3.1, Histone acetyltransferase KAT6A, ... | | Authors: | Qiu, Y, Li, F. | | Deposit date: | 2011-12-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription
Genes Dev., 26, 2012
|
|
8ZQO
 
 | | Human high-affinity choline transporter CHT1 bound to HC-3 under NaCl condition, with sodium and chloride ions coordinated. | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), CHLORIDE ION, High affinity choline transporter 1, ... | | Authors: | Qiu, Y, Gao, Y, Zhao, Y. | | Deposit date: | 2024-06-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Ion coupling and inhibitory mechanisms of the human presynaptic high-affinity choline transporter CHT1.
Structure, 33, 2025
|
|
8ZQR
 
 | | Human high-affinity choline transporter CHT1 in apo state under NaCl condition. | | Descriptor: | High affinity choline transporter 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Qiu, Y, Gao, Y, Zhao, Y. | | Deposit date: | 2024-06-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ion coupling and inhibitory mechanisms of the human presynaptic high-affinity choline transporter CHT1.
Structure, 33, 2025
|
|
8ZQP
 
 | | Human high-affinity choline transporter CHT1 bound to ML352 under NaCl condition, with sodium ion coordinated. | | Descriptor: | 4-methoxy-3-[(1-methylpiperidin-4-yl)oxy]-N-{[3-(propan-2-yl)-1,2-oxazol-5-yl]methyl}benzamide, High affinity choline transporter 1, SODIUM ION, ... | | Authors: | Qiu, Y, Gao, Y, Zhao, Y. | | Deposit date: | 2024-06-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ion coupling and inhibitory mechanisms of the human presynaptic high-affinity choline transporter CHT1.
Structure, 33, 2025
|
|
8ZQQ
 
 | | Human high-affinity choline transporter CHT1 bound to choline under NaCl condition, with sodium and chloride ions coordinated. | | Descriptor: | CHLORIDE ION, CHOLINE ION, High affinity choline transporter 1, ... | | Authors: | Qiu, Y, Gao, Y, Zhao, Y. | | Deposit date: | 2024-06-03 | | Release date: | 2024-12-04 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Ion coupling and inhibitory mechanisms of the human presynaptic high-affinity choline transporter CHT1.
Structure, 33, 2025
|
|
6PDX
 
 | |
5D7G
 
 | |
6XGC
 
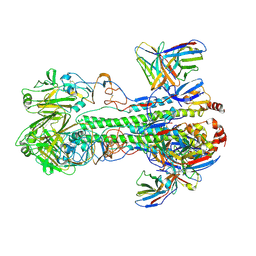 | |
6WZT
 
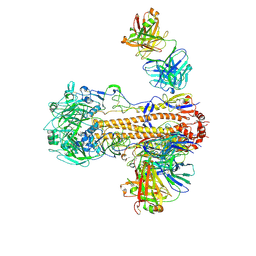 | | CryoEM structure of influenza hemagglutinin A/Victoria/361/2011 in complex with cyno antibody 3B10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cyno antibody heavy chain, ... | | Authors: | Qiu, Y, Zhou, Y, Darricarrere, N. | | Deposit date: | 2020-05-14 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Broad neutralization of H1 and H3 viruses by adjuvanted influenza HA stem vaccines in nonhuman primates.
Sci Transl Med, 13, 2021
|
|
9BDO
 
 | | Crystal structure of anti-abTCR NANOBODY VHH | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Qiu, Y. | | Deposit date: | 2024-04-12 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Identification and non-clinical characterization of SAR444200, a novel anti-GPC3 NANOBODY T cell engager, for the treatment of GPC3+ solid tumors
To Be Published
|
|
2HV2
 
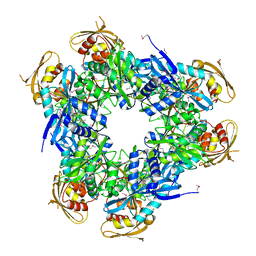 | | Crystal Structure of Conserved Protein of Unknown Function from Enterococcus faecalis V583 at 2.4 A Resolution, Probable N-Acyltransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein, TETRAETHYLENE GLYCOL | | Authors: | Tereshko, V.A, Qiu, Y, Kossiakoff, A.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-27 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Enterococcus faecalis V583 at 2.4 A resolution.
To be Published
|
|
8J77
 
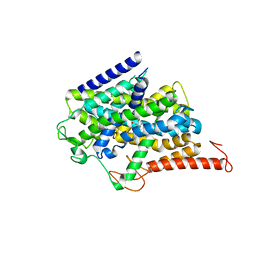 | |
8J75
 
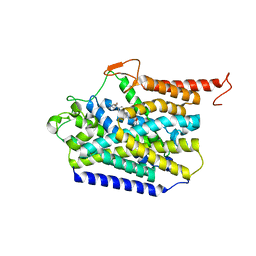 | | Human high-affinity choline transporter CHT1 in the HC-3-bound outward-facing open conformation, monomeric state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), High affinity choline transporter 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gao, Y, Qiu, Y, Zhao, Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transport mechanism of presynaptic high-affinity choline uptake by CHT1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J76
 
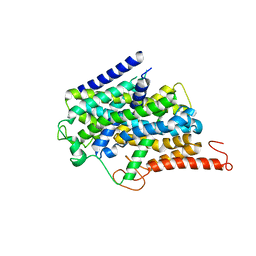 | |
8J74
 
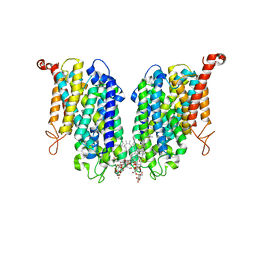 | | Human high-affinity choline transporter CHT1 in the HC-3-bound outward-facing open conformation, dimeric state | | Descriptor: | (2S,2'S)-2,2'-biphenyl-4,4'-diylbis(2-hydroxy-4,4-dimethylmorpholin-4-ium), CHOLESTEROL HEMISUCCINATE, HEXADECANE, ... | | Authors: | Gao, Y, Qiu, Y, Zhao, Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transport mechanism of presynaptic high-affinity choline uptake by CHT1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1T34
 
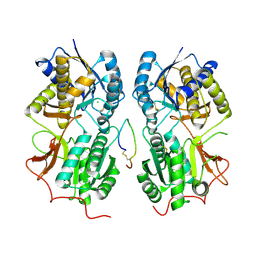 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | Authors: | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | Deposit date: | 2004-04-23 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
7XLQ
 
 | | Structure of human R-type voltage-gated CaV2.3-alpha2/delta1-beta1 channel complex in the ligand-free (apo) state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gao, Y, Qiu, Y, Wei, Y, Dong, Y, Zhang, X.C, Zhao, Y. | | Deposit date: | 2022-04-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into the gating mechanisms of voltage-gated calcium channel Ca V 2.3.
Nat Commun, 14, 2023
|
|
