2AB6
 
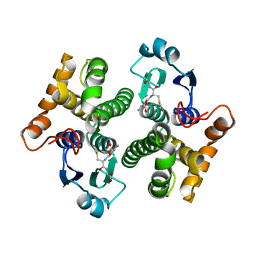 | |
4MF5
 
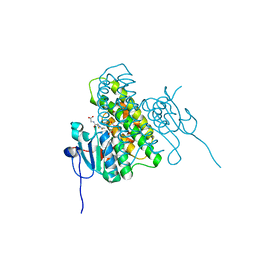 | | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Crystal structure of glutathione transferase BgramDRAFT_1843 from Burkholderia graminis, Target EFI-507289, with traces of one GSH bound
To be Published
|
|
4MZY
 
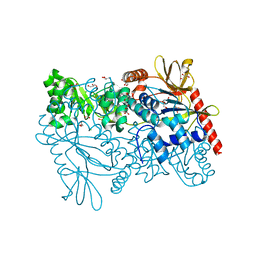 | | Crystal structure of enterococcus faecalis nicotinate phosphoribosyltransferase with malonate and phosphate bound | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Wasserman, S.R, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC), New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Enterococcus Faecalis Nicotinate Phosphoribosyltransferase
To be Published
|
|
2Q01
 
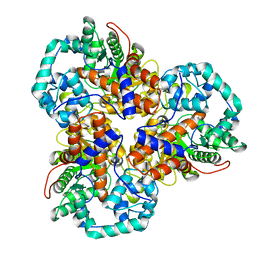 | | Crystal structure of glucuronate isomerase from Caulobacter crescentus | | Descriptor: | POTASSIUM ION, Uronate isomerase | | Authors: | Patskovsky, Y, Bonanno, J, Sridhar, V, Sauder, J.M, Freeman, J, Powell, A, Koss, J, Groshong, C, Gheyi, T, Wasserman, S.R, Raushel, F, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-18 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Glucuronate Isomerase from Caulobacter crescentus.
To be Published
|
|
2AA4
 
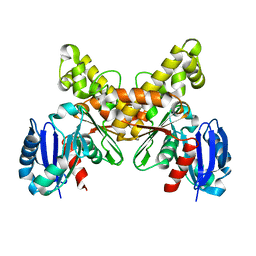 | |
7N9J
 
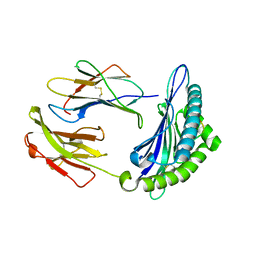 | | Crystal structure of H2DB in complex with HSF2 melanoma neoantigen | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the complex between H2DB and melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
7NA5
 
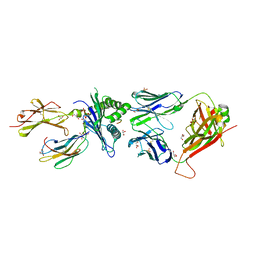 | | Structure of the H2DB-TCR ternary complex with HSF2 melanoma neoantigen | | Descriptor: | 47BE7 TCR alpha chain, 47BE7 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Patskovsky, Y, Finnigan, J, Patskovska, L, Newman, J, Bhardwaj, N, Krogsgaard, M. | | Deposit date: | 2021-06-19 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the TCR-H2DB ternary complex with melanoma HSF2 neoantigen YGFRNVVHI
To be Published
|
|
2B0A
 
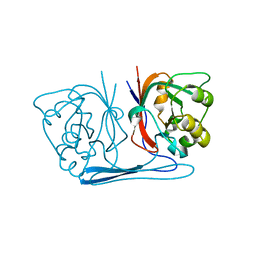 | |
4DFD
 
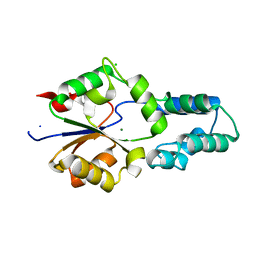 | | Crystal structure of had family enzyme bt-2542 (target efi-501088) from bacteroides thetaiotaomicron, magnesium complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative haloacid dehalogenase-like hydrolase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-23 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
4DCC
 
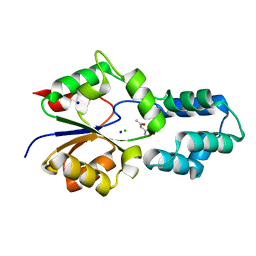 | | Crystal structure of had family enzyme bt-2542 from bacteroides thetaiotaomicron (target efi-501088) | | Descriptor: | CHLORIDE ION, Putative haloacid dehalogenase-like hydrolase, SODIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Allen, K.N, Dunaway-Mariano, D, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of protein Bt-2542 from Bacteroides Thetaiotaomicron (Target Efi-501088)
To be Published
|
|
3KEW
 
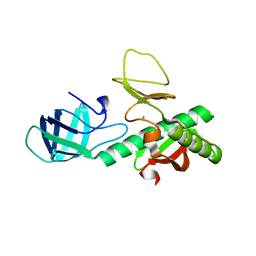 | | Crystal structure of probable alanyl-trna-synthase from Clostridium perfringens | | Descriptor: | DHHA1 domain protein, ZINC ION | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alanyl-trna-synthase from Clostridium perfringens
To be Published
|
|
3KSU
 
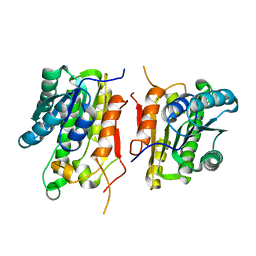 | | Crystal structure of short-chain dehydrogenase from oenococcus oeni psu-1 | | Descriptor: | 3-oxoacyl-acyl carrier protein reductase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-11-23 | | Release date: | 2009-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase from Oenococcus Oeni Psu-1
To be Published
|
|
1S7J
 
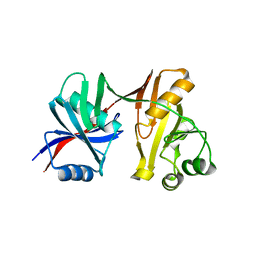 | |
1TE5
 
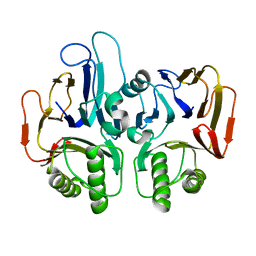 | |
1XW6
 
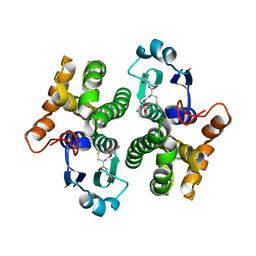 | | 1.9 angstrom resolution structure of human glutathione S-transferase M1A-1A complexed with glutathione | | Descriptor: | GLUTATHIONE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-10-29 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
1XWK
 
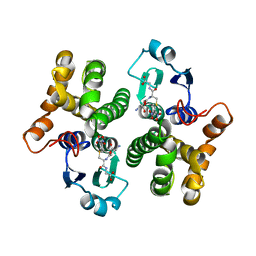 | | 2.3 angstrom resolution crystal structure of human glutathione S-transferase M1A-1A complexed with glutathionyl-S-dinitrobenzene | | Descriptor: | GLUTATHIONE S-(2,4 DINITROBENZENE), Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2004-11-01 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
3BH1
 
 | | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae | | Descriptor: | GLYCEROL, UPF0371 protein DIP2346 | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Gilmore, M, Iizuka, M, Groshong, C, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of protein DIP2346 from Corynebacterium diphtheriae.
To be Published
|
|
3BJS
 
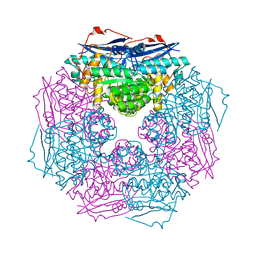 | | Crystal structure of a member of enolase superfamily from Polaromonas sp. JS666 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme | | Authors: | Patskovsky, Y, Bonanno, J.B, Ozyurt, S, Dickey, M, Sauder, J.M, Reyes, C, Groshong, C, Gheyi, T, Smith, D, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Member of Enolase Superfamily from Polaromonas sp. JS666.
To be Published
|
|
3B5M
 
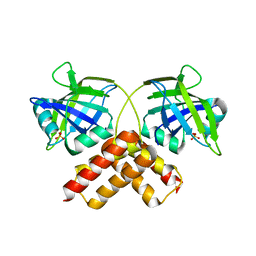 | | Crystal structure of conserved uncharacterized protein from Rhodopirellula baltica | | Descriptor: | SULFATE ION, Uncharacterized protein | | Authors: | Patskovsky, Y, Bonanno, J.B, Sridhar, V, Rutter, M, Powell, A, Maletic, M, Rodgers, R, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-26 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Conserved Uncharacterized Protein from Rhodopirellula baltica.
To be Published
|
|
2F3M
 
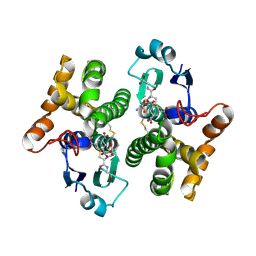 | | Structure of human GLUTATHIONE S-TRANSFERASE M1A-1A complexed with 1-(S-(GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXADIENATE ANION | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, Glutathione S-transferase Mu 1 | | Authors: | Patskovsky, Y, Patskovska, L, Almo, S.C, Listowsky, I. | | Deposit date: | 2005-11-21 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Transition state model and mechanism of nucleophilic aromatic substitution reactions catalyzed by human glutathione S-transferase M1a-1a.
Biochemistry, 45, 2006
|
|
7RZD
 
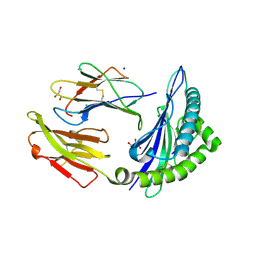 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8I
 
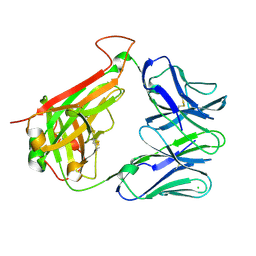 | | PHOSPHOPEPTIDE-SPECIFIC LC13 TCR, MONOCLINIC CRYSTAL FORM | | Descriptor: | CHLORIDE ION, TRAV27_LC13 TCR ALPHA CHAIN, TRBV27_LC13 TCR BETA CHAIN | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8E
 
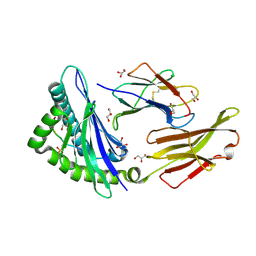 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8F
 
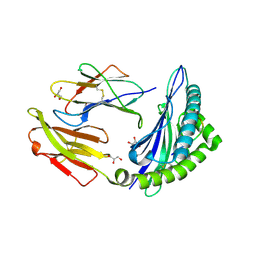 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
