7AA9
 
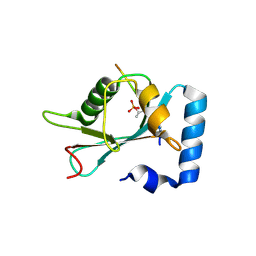 | | Structure of SCOC pT13/pT15 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pT13/PT15 SCOC LIR | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA8
 
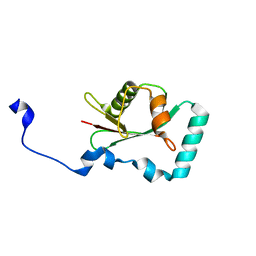 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA7
 
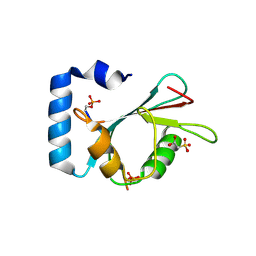 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
6HOL
 
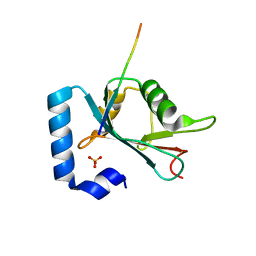 | | Structure of ATG14 LIR motif bound to GABARAPL1 | | Descriptor: | Beclin 1-associated autophagy-related key regulator, GLYCEROL, Gamma-aminobutyric acid receptor-associated protein-like 1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HYL
 
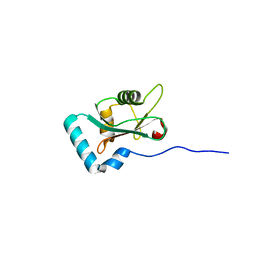 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.559 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HOH
 
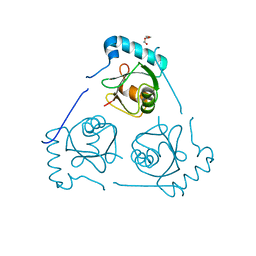 | | Structure of VPS34 LIR motif (S249E) bound to GABARAP | | Descriptor: | Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, TRIETHYLENE GLYCOL | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOK
 
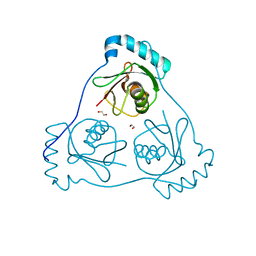 | | Structure of Beclin1 LIR (S96E) motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOG
 
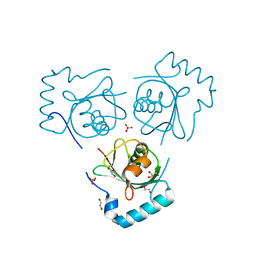 | | Structure of VPS34 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3,Gamma-aminobutyric acid receptor-associated protein, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HOJ
 
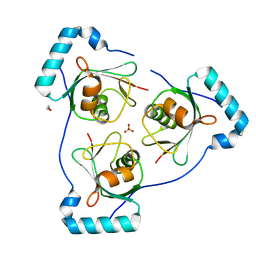 | | Structure of Beclin1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, Beclin-1,Gamma-aminobutyric acid receptor-associated protein, SULFATE ION | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HYO
 
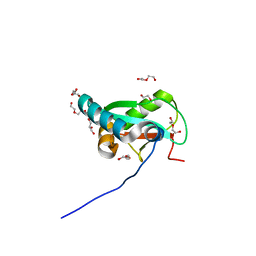 | | Structure of ULK1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HYN
 
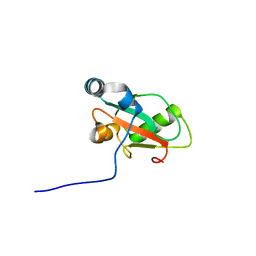 | | Structure of ATG13 LIR motif bound to GABARAP | | Descriptor: | Autophagy-related protein 13,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
6HOI
 
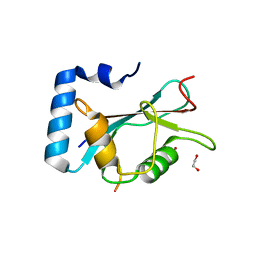 | | Structure of Beclin1 LIR motif bound to GABARAPL1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beclin-1, ... | | Authors: | Mouilleron, S, Birgisdottir, A.B, Bhujbal, Z, Wirth, M, Sjottem, E, Evjen, G, Zhang, W, Lee, R, O'Reilly, N, Tooze, S, Lamark, T, Johansen, T. | | Deposit date: | 2018-09-17 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs.
Autophagy, 15, 2019
|
|
6HYM
 
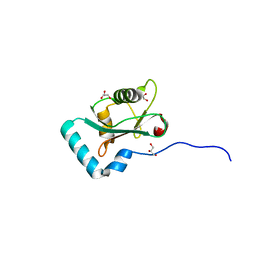 | | Structure of PCM1 LIR motif bound to GABARAP | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Pericentriolar material 1 protein,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Mouilleron, S, Wirth, M, Zhang, W, O'Reilly, N, Tooze, S, Johansen, T, Razi, M, Nyoni, L, Joshi, D. | | Deposit date: | 2018-10-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins.
Nat Commun, 10, 2019
|
|
4BG6
 
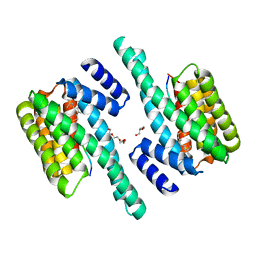 | | 14-3-3 interaction with Rnd3 prenyl-phosphorylation motif | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FARNESYL, ... | | Authors: | Riou, P, Kjaer, S, Purkiss, A, O'Reilly, N, McDonald, N.Q. | | Deposit date: | 2013-03-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 14-3-3 Proteins Interact with a Hybrid Prenyl-Phosphorylation Motif to Inhibit G Proteins.
Cell(Cambridge,Mass.), 153, 2013
|
|
4B1Y
 
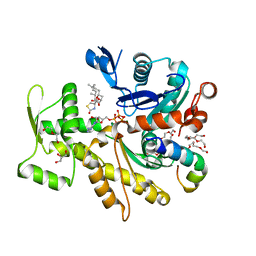 | | Structure of the Phactr1 RPEL-3 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1X
 
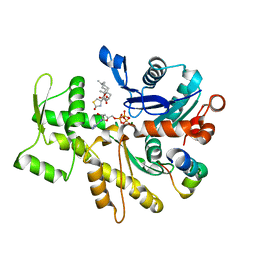 | | Structure of the Phactr1 RPEL-2 bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1U
 
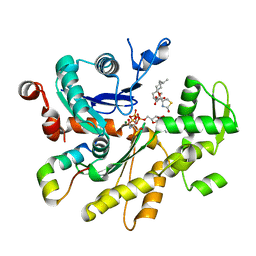 | | Structure of the Phactr1 RPEL domain and RPEL motif directed assemblies with G-actin reveal the molecular basis for actin binding cooperativity. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1Z
 
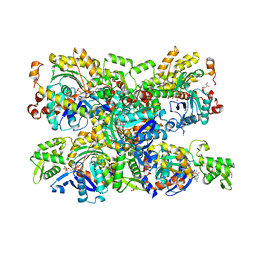 | | Structure of the Phactr1 RPEL domain bound to G-actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2012-11-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1V
 
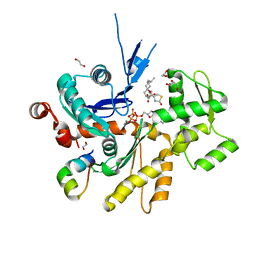 | | Structure of the Phactr1 RPEL-N domain bound to G-actin | | Descriptor: | 1,2-ETHANEDIOL, ACTIN, ALPHA SKELETAL MUSCLE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
4B1W
 
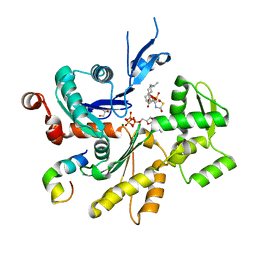 | | Structure of the Phactr1 RPEL-2 domain bound to actin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mouilleron, S, Wiezlak, M, O'Reilly, N, Treisman, R, McDonald, N.Q. | | Deposit date: | 2012-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the Phactr1 RPEL domain and RPEL motif complexes with G-actin reveal the molecular basis for actin binding cooperativity.
Structure, 20, 2012
|
|
6ZEH
 
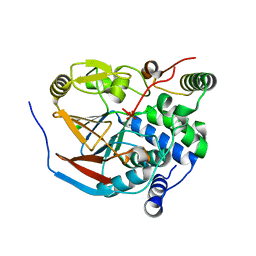 | | Structure of PP1-spectrin alpha II chimera [PP1(7-304) + linker (G/S)x9 + spectrin alpha II (1025-1039)] bound to Phactr1 (516-580) | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Phosphatase and actin regulator, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEI
 
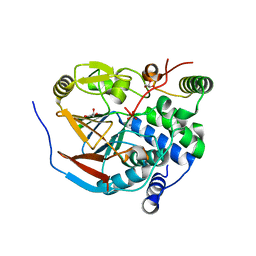 | | Structure of PP1-IRSp53 S455E chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEE
 
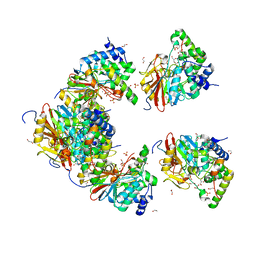 | | Structure of PP1(7-300) bound to Phactr1 (507-580) at pH8.4 | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, GLYCEROL, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEF
 
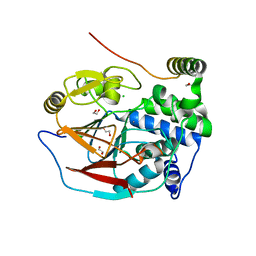 | | Structure of PP1(7-300) bound to Phactr1 (516-580) at pH 5.25 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
6ZEG
 
 | | Structure of PP1-IRSp53 chimera [PP1(7-304) + linker (G/S)x9 + IRSp53(449-465)] bound to Phactr1 (516-580) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE, MANGANESE (II) ION, ... | | Authors: | Mouilleron, S, Treisman, R, Fedoryshchak, R, Lee, R, Butler, A.M, Prechova, M. | | Deposit date: | 2020-06-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Molecular basis for substrate specificity of the Phactr1/PP1 phosphatase holoenzyme.
Elife, 9, 2020
|
|
