4R5Z
 
 | | Crystal structure of Rv3772 encoded aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
4RAE
 
 | |
4R2N
 
 | | Crystal structure of Rv3772 in complex with its substrate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
4R8D
 
 | | Crystal structure of Rv1600 encoded aminotransferase in complex with PLP-MES from Mycobacterium tuberculosis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Histidinol-phosphate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-09-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Rv1600 encoded aminotransferase in complex with PLP-MES from Mycobacterium tuberculosis
To be Published
|
|
8PEF
 
 | |
6S73
 
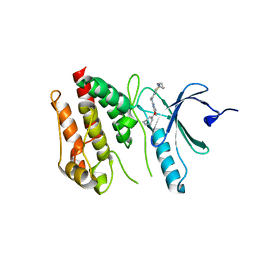 | | Crystal structure of Nek7 SRS mutant bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Byrne, M.J, Bhatia, C, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
6S75
 
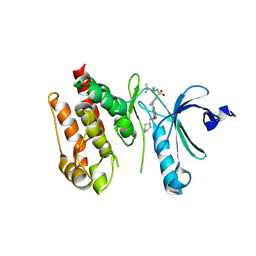 | | Crystal structure of Nek7 bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
6S76
 
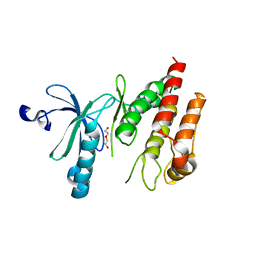 | | Crystal structure of human Nek7 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
4GQU
 
 | | Crystal structure of HisB from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ahangar, M.S, Vyas, R, Nasir, N, Biswal, B.K. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of the native, substrate-
bound and inhibited forms of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase
Acta Crystallogr.,Sect.D, 2013
|
|
4LOM
 
 | | Crystal Structure of Mycobacterium tuberculosis HisB in complex with its substrate | | Descriptor: | (2R,3S)-2,3-dihydroxy-3-(1H-imidazol-5-yl)propyl dihydrogen phosphate, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ahangar, M.S, Vyas, R, Nasir, N, Biswal, B.K. | | Deposit date: | 2013-07-13 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the native, substrate-
bound and inhibited forms of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase
Acta Crystallogr.,Sect.D, 2013
|
|
4LPF
 
 | | Crystal structure of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in complex with an inhibitor | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Ahangar, M.S, Vyas, R, Nasir, N, Biswal, B.K. | | Deposit date: | 2013-07-16 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the native, substrate-
bound and inhibited forms of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase
Acta Crystallogr.,Sect.D, 2013
|
|
6SK9
 
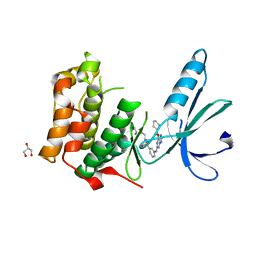 | | Nek2 bound to purine compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, GLYCEROL, Serine/threonine-protein kinase Nek2 | | Authors: | Bayliss, R, Byrne, M.J, Mas-Droux, C. | | Deposit date: | 2019-08-15 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
