2GAQ
 
 | |
1RQM
 
 | | SOLUTION STRUCTURE OF THE K18G/R82E ALICYCLOBACILLUS ACIDOCALDARIUS THIOREDOXIN MUTANT | | Descriptor: | Thioredoxin | | Authors: | Leone, M, Di Lello, P, Ohlenschlager, O, Pedone, E.M, Bartolucci, S, Rossi, M, Di Blasio, B, Pedone, C, Saviano, M, Isernia, C, Fattorusso, R. | | Deposit date: | 2003-12-05 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the K18G/R82E Alicyclobacillus acidocaldarius Thioredoxin Mutant: A Molecular Analysis of Its Reduced Thermal Stability.
Biochemistry, 43, 2004
|
|
2K4P
 
 | | Solution Structure of Ship2-Sam | | Descriptor: | Phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Leone, M, Pellecchia, M. | | Deposit date: | 2008-06-16 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of a Heterotypic Sam-Sam Domain Association: The Interaction between the Lipid Phosphatase Ship2 and the EphA2 Receptor.
Biochemistry, 47, 2008
|
|
2KG5
 
 | | NMR Solution structure of ARAP3-SAM | | Descriptor: | Arf-GAP, Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Leone, M, Pellecchia, M. | | Deposit date: | 2009-03-05 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Sam domain of the lipid phosphatase Ship2 adopts a common model to interact with Arap3-Sam and EphA2-Sam.
Bmc Struct.Biol., 9, 2009
|
|
2GJY
 
 | |
2LMR
 
 | | Solution structure of the first sam domain of odin | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 1A | | Authors: | Leone, M, Mercurio, F. | | Deposit date: | 2011-12-12 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the First Sam Domain of Odin and Binding Studies with the EphA2 Receptor.
Biochemistry, 51, 2012
|
|
6F7O
 
 | | NMR structure of an Odin-Sam1 stapled peptide | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 1A | | Authors: | Leone, M, Mercurio, F.A. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6FS4
 
 | |
1NJQ
 
 | | NMR structure of the single QALGGH zinc finger domain from Arabidopsis thaliana SUPERMAN protein | | Descriptor: | ZINC ION, superman protein | | Authors: | Isernia, C, Bucci, E, Leone, M, Zaccaro, L, Di Lello, P, Digilio, G, Esposito, S, Saviano, M, Di Blasio, B, Pedone, C, Pedone, P.V, Fattorusso, R. | | Deposit date: | 2003-01-02 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Single QALGGH Zinc Finger Domain from the Arabidopsis thaliana SUPERMAN Protein.
Chembiochem, 4, 2003
|
|
2MYQ
 
 | | NMR structure of an Odin-Sam1 fragment | | Descriptor: | Ankyrin repeat and SAM domain-containing protein 1A | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2015-01-30 | | Release date: | 2015-07-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Peptide Fragments of Odin-Sam1: Conformational Analysis and Interaction Studies with EphA2-Sam.
Chembiochem, 16, 2015
|
|
2A25
 
 | | Crystal structure of Siah1 SBD bound to the peptide EKPAAVVAPITTG from SIP | | Descriptor: | Calcyclin-binding protein peptide, Ubiquitin ligase SIAH1, ZINC ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
2A26
 
 | | Crystal structure of the N-terminal, dimerization domain of Siah Interacting Protein | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Calcyclin-binding protein, SULFATE ION | | Authors: | Santelli, E, Leone, M, Li, C, Fukushima, T, Preece, N.E, Olson, A.J, Ely, K.R, Reed, J.C, Pellecchia, M, Liddington, R.C, Matsuzawa, S. | | Deposit date: | 2005-06-21 | | Release date: | 2005-08-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Analysis of Siah1-Siah-interacting Protein Interactions and Insights into the Assembly of an E3 Ligase Multiprotein Complex
J.Biol.Chem., 280, 2005
|
|
5NZ9
 
 | | NMR structure of an EphA2-Sam fragment | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-05-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The Sam-Sam interaction between Ship2 and the EphA2 receptor: design and analysis of peptide inhibitors.
Sci Rep, 7, 2017
|
|
2L1O
 
 | | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys2His2 zinc finger induces structural rearrangements of typical DNA base determinant positions | | Descriptor: | CADMIUM ION, Transcriptional regulator SUPERMAN | | Authors: | Malgieri, G, Zaccaro, L, Leone, M, Bucci, E, Esposito, S, Baglivo, I, Del Gatto, A, Scandurra, R, Pedone, P.V, Fattorusso, R, Isernia, C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys(2) His(2) zinc finger induces structural rearrangements of typical DNA base determinant positions.
Biopolymers, 95, 2011
|
|
6F7M
 
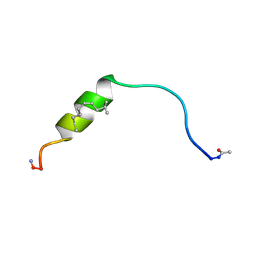 | | NMR structure of EphA2-Sam stapled peptides (S13ST) | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6F7N
 
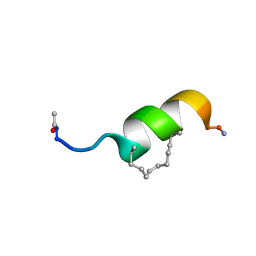 | | NMR structure of EphA2-Sam stapled peptides (S13STshort) | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Mercurio, F.A, Leone, M. | | Deposit date: | 2017-12-11 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Sam domain-based stapled peptides: Structural analysis and interaction studies with the Sam domains from the EphA2 receptor and the lipid phosphatase Ship2.
Bioorg. Chem., 80, 2018
|
|
6FS5
 
 | |
2IH1
 
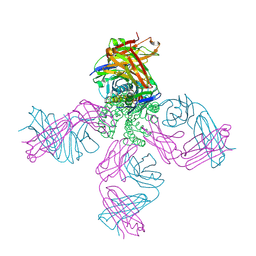 | | Ion selectivity in a semi-synthetic K+ channel locked in the conductive conformation | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB Heavy Chain, FAB Light Chain, ... | | Authors: | Valiyaveetil, F.I, Leonetti, M, Muir, T.W, MacKinnon, R. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ion Selectivity in a Semisynthetic K+ Channel Locked in the Conductive Conformation.
Science, 314, 2006
|
|
2IH3
 
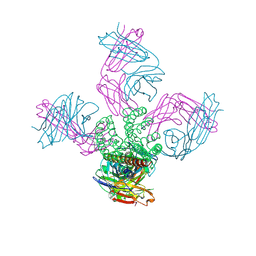 | | Ion selectivity in a semi-synthetic K+ channel locked in the conductive conformation | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, FAB Heavy Chain, FAB Light Chain, ... | | Authors: | Valiyaveetil, F.I, Leonetti, M, Muir, T.W, MacKinnon, R. | | Deposit date: | 2006-09-25 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ion Selectivity in a Semisynthetic K+ Channel Locked in the Conductive Conformation.
Science, 314, 2006
|
|
1UWI
 
 | |
5NHU
 
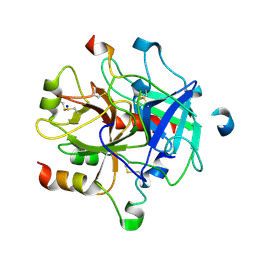 | |
2EXJ
 
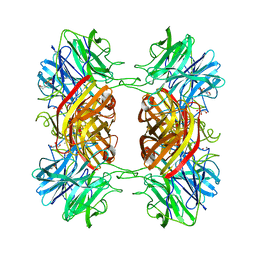 | | Structure of the family43 beta-Xylosidase D128G mutant from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXI
 
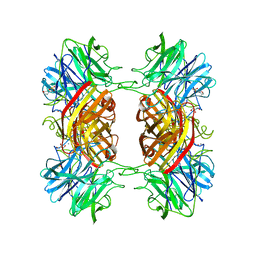 | | Structure of the family43 beta-Xylosidase D15G mutant from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXK
 
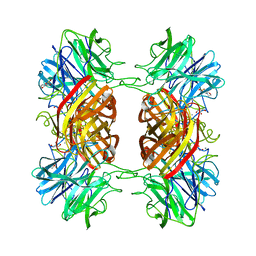 | | Structure of the family43 beta-Xylosidase E187G from geobacillus stearothermophilus in complex with xylobiose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Shoham, Y, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
2EXH
 
 | | Structure of the family43 beta-Xylosidase from geobacillus stearothermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Brux, C, Niefind, K, Shallom-Shezifi, D, Yuval, S, Schomburg, D. | | Deposit date: | 2005-11-08 | | Release date: | 2006-04-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Structure of an Inverting GH43 beta-Xylosidase from Geobacillus stearothermophilus with its Substrate Reveals the Role of the Three Catalytic Residues.
J.Mol.Biol., 359, 2006
|
|
