7WG3
 
 | | Structural basis of interleukin-17B receptor in complex with a neutralizing antibody D9 for guiding humanization and affinity maturation for cancer therapy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D9 Fab, IL17RB protein, ... | | Authors: | Lee, W.H, Chen, X.R, Liu, I.J, Lee, J.H, Hu, C.M, Wu, H.C, Wang, S.K, Lee, W.H, Ma, C. | | Deposit date: | 2021-12-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of interleukin-17B receptor in complex with a neutralizing antibody for guiding humanization and affinity maturation.
Cell Rep, 41, 2022
|
|
1L9W
 
 | | CRYSTAL STRUCTURE OF 3-DEHYDROQUINASE FROM SALMONELLA TYPHI COMPLEXED WITH REACTION PRODUCT | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase aroD | | Authors: | Lee, W.H, Perles, L.A, Nagem, R.A.P, Shrive, A.K, Hawkins, A, Sawyer, L, Polikarpov, I. | | Deposit date: | 2002-03-26 | | Release date: | 2003-03-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comparison of different crystal forms of 3-dehydroquinase from Salmonella typhi and its implication for the enzyme activity.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3NGL
 
 | | Crystal structure of bifunctional 5,10-methylenetetrahydrofolate dehydrogenase / cyclohydrolase from Thermoplasma acidophilum | | Descriptor: | Bifunctional protein folD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sung, M.W, Lee, W.H, Hwang, K.Y. | | Deposit date: | 2010-06-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of bifunctional 5,10-methylenetetrahydrofolate dehydrogenase/cyclohydrolase from Thermoplasma acidophilum
Biochem.Biophys.Res.Commun., 406, 2011
|
|
3NGX
 
 | |
1MIU
 
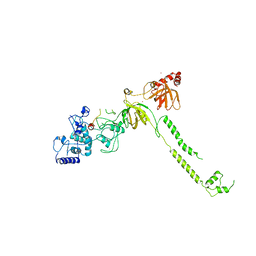 | | Structure of a BRCA2-DSS1 complex | | Descriptor: | Breast Cancer type 2 susceptibility protein, Deleted in split hand/split foot protein 1, MERCURY (II) ION | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA
structure
Science, 297, 2002
|
|
8U53
 
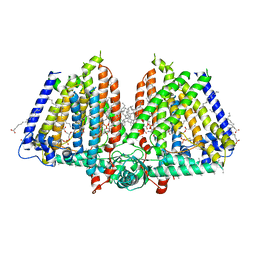 | | Mechanically activated ion channel OSCA3.1 in nanodiscs | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CSC1-like protein ERD4, PALMITIC ACID, ... | | Authors: | Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-09-12 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided mutagenesis of OSCAs reveals differential activation to mechanical stimuli.
Elife, 12, 2024
|
|
8TQ1
 
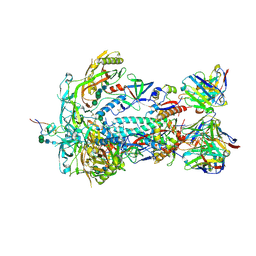 | | HIV-1 BG505 Env SOSIP in complex with bovine Fab Bess4 and non-human primate Fab RM20A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Bovine Bess4 Fab heavy chain, ... | | Authors: | Ozorowski, G, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire.
Plos Pathog., 20, 2024
|
|
1MJE
 
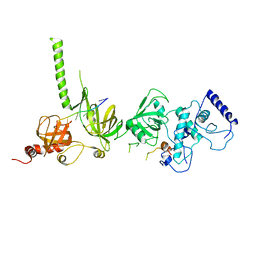 | | STRUCTURE OF A BRCA2-DSS1-SSDNA COMPLEX | | Descriptor: | 5'-D(P*TP*TP*TP*TP*TP*T)-3', Deleted in split hand/split foot protein 1, breast cancer 2 | | Authors: | Yang, H, Jeffrey, P.D, Miller, J, Kinnucan, E, Sun, Y, Thoma, N.H, Zheng, N, Chen, P.L, Lee, W.H, Pavletich, N.P. | | Deposit date: | 2002-08-27 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure.
Science, 297, 2002
|
|
8T57
 
 | | Structure of mechanically activated ion channel OSCA2.3 in peptidiscs | | Descriptor: | CHOLESTEROL, CSC1-like protein HYP1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Burendei, B, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
8T56
 
 | | Structure of mechanically activated ion channel OSCA1.2 in peptidiscs | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Burendei, B, Jojoa-Cruz, S, Lee, W.H, Ward, A.B. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of mechanically activated ion channel OSCA2.3 reveals mobile elements in the transmembrane domain.
Structure, 32, 2024
|
|
6NF4
 
 | | Structure of zebrafish Otop1 in nanodiscs | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, Otopetrin1 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NF6
 
 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2OHE
 
 | | Structural and mutational analysis of tRNA-Intron splicing endonuclease from Thermoplasma acidophilum DSM 1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
2OHC
 
 | | structural and mutational analysis of tRNA-intron splicing endonuclease from Thermoplasma acidophilum DSM1728 | | Descriptor: | tRNA-splicing endonuclease | | Authors: | Kim, Y.K, Mizutani, K, Rhee, K.H, Lee, W.H, Park, S.Y, Hwang, K.Y. | | Deposit date: | 2007-01-10 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Mutational Analysis of tRNA Intron-Splicing Endonuclease from Thermoplasma acidophilum DSM 1728: Catalytic Mechanism of tRNA Intron-Splicing Endonucleases
J.Bacteriol., 189, 2007
|
|
3KKL
 
 | |
7N5F
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Down' conformation in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5D
 
 | | Composite Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5G
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 Protomer in 'Up' conformation in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
7N5E
 
 | | Structure of Mechanosensitive Ion Channel Flycatcher1 in GDN | | Descriptor: | Mechanosensitive ion channel Flycatcher1, PALMITIC ACID | | Authors: | Jojoa-Cruz, S, Saotome, K, Lee, W.H, Patapoutian, A, Ward, A.B. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the Venus flytrap mechanosensitive ion channel Flycatcher1.
Nat Commun, 13, 2022
|
|
1QLL
 
 | | Piratoxin-II (Prtx-II) - a K49 PLA2 from Bothrops pirajai | | Descriptor: | N-TRIDECANOIC ACID, PHOSPHOLIPASE A2 | | Authors: | Lee, W.-H, Polikarpov, I. | | Deposit date: | 1999-09-01 | | Release date: | 2000-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Low Catalytic Activity in Lys49 Phospholipases A2-A Hypothesis: The Crystal Structure of Piratoxin II Complexed to Fatty Acid
Biochemistry, 40, 2001
|
|
2QEP
 
 | | Crystal structure of the D1 domain of PTPRN2 (IA2beta) | | Descriptor: | CHLORIDE ION, Receptor-type tyrosine-protein phosphatase N2 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Berridge, G, Burgess-Brown, N, Das, S, Fedorov, O, King, O, Niesen, F, Watt, S, Savitsky, P, Salah, E, Pike, A.C.W, Bunkoczi, G, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-26 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2F57
 
 | | Crystal Structure Of The Human P21-Activated Kinase 5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N2-[(1R,2S)-2-AMINOCYCLOHEXYL]-N6-(3-CHLOROPHENYL)-9-ETHYL-9H-PURINE-2,6-DIAMINE, Serine/threonine-protein kinase PAK 7 | | Authors: | Eswaran, J, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Bray, J, Das, S, Savitsky, P, Smee, C, Burgess, N, Fedorov, O, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-25 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of the p21-activated kinases PAK4, PAK5, and PAK6 reveal catalytic domain plasticity of active group II PAKs.
Structure, 15, 2007
|
|
7OVZ
 
 | |
6DJB
 
 | | Structure of human Volume Regulated Anion Channel composed of SWELL1 (LRRC8A) | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kefauver, J.M, Saotome, K, Pallesen, J, Cottrell, C.A, Ward, A.B, Patapoutian, A. | | Deposit date: | 2018-05-24 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the human volume regulated anion channel.
Elife, 7, 2018
|
|
8TGO
 
 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
