5MM2
 
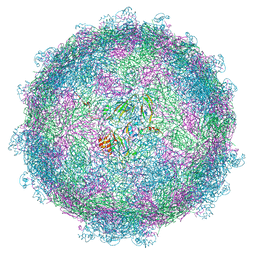 | | nora virus structure | | Descriptor: | Capsid protein VP4A, capsid protein VP4B, capsid protein VP4C | | Authors: | Laurinmaki, P, Shakeel, S, Ekstrom, J.-O, Butcher, S.J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of Nora virus at 2.7 angstrom resolution and implications for receptor binding, capsid stability and taxonomy.
Sci Rep, 10, 2020
|
|
7ZFR
 
 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide bound in the reverse direction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II HLA-DP alpha chain (DPA1*02:01), MHC class II HLA-DP beta chain (DPB1*01:01), ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-04-01 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
7ZAK
 
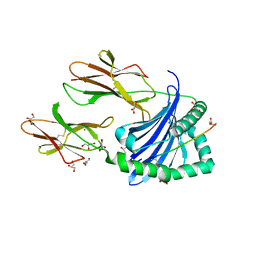 | | Crystal structure of HLA-DP (DPA1*02:01-DPB1*01:01) in complex with a peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Racle, J, Guillaume, P, Larabi, A, Lau, K, Pojer, F, Gfeller, D. | | Deposit date: | 2022-03-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Machine learning predictions of MHC-II specificities reveal alternative binding mode of class II epitopes.
Immunity, 56, 2023
|
|
4GAG
 
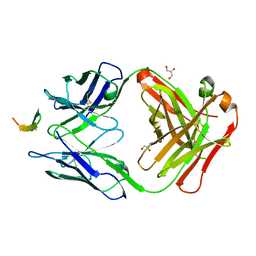 | | Structure of the broadly neutralizing antibody AP33 in complex with its HCV epitope (E2 residues 412-423) | | Descriptor: | GLYCEROL, Genome polyprotein, NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, ... | | Authors: | Potter, J.A, Owsianka, A, Angus, A.G.N, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-25 | | Release date: | 2012-10-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
4GAY
 
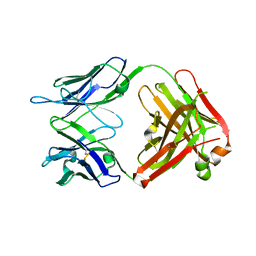 | | Structure of the broadly neutralizing antibody AP33 | | Descriptor: | NEUTRALIZING ANTIBODY AP33 HEAVY CHAIN, NEUTRALIZING ANTIBODY AP33 LIGHT CHAIN, TRIETHYLENE GLYCOL | | Authors: | Potter, J.A, Owsianka, A, Jeffery, N, Matthews, D, Keck, Z, Lau, P, Foung, S.K.H, Taylor, G.L, Patel, A.H. | | Deposit date: | 2012-07-26 | | Release date: | 2012-10-10 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Toward a Hepatitis C Virus Vaccine: the Structural Basis of Hepatitis C Virus Neutralization by AP33, a Broadly Neutralizing Antibody.
J.Virol., 86, 2012
|
|
8KD0
 
 | |
8WCH
 
 | |
8X2V
 
 | |
8X2W
 
 | |
8HNE
 
 | | Crystal structure of the ancestral GH19 chitinase Anc4 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Anc4, ... | | Authors: | Kozome, D, Laurino, P. | | Deposit date: | 2022-12-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Remote loop evolution reveals a complex biological function for chitinase enzymes beyond the active site.
Nat Commun, 15, 2024
|
|
8HNF
 
 | | Crystal structure of the ancestral GH19 chitinase Anc5 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Anc5, ... | | Authors: | Kozome, D, Laurino, P. | | Deposit date: | 2022-12-07 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Remote loop evolution reveals a complex biological function for chitinase enzymes beyond the active site.
Nat Commun, 15, 2024
|
|
8HQQ
 
 | |
8HQR
 
 | |
3J2J
 
 | | Empty coxsackievirus A9 capsid | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Shakeel, S, Seitsonen, J.J.T, Kajander, T, Laurinmaki, P, Hyypia, T, Susi, P, Butcher, S.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.54 Å) | | Cite: | Structural and functional analysis of coxsackievirus A9 integrin {alpha}v{beta}6 binding and uncoating.
J.Virol., 87, 2013
|
|
6EI2
 
 | | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CADMIUM ION, ... | | Authors: | Picaud, S, Guillaume, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gfeller, D, Filippakopoulos, P. | | Deposit date: | 2017-09-16 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of HLA-A68 presenting a C-terminally extended peptide
To Be Published
|
|
6WFS
 
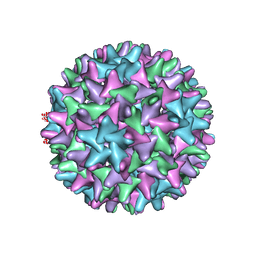 | | Cryo-EM Structure of Hepatitis B virus T=4 capsid in complex with the antiviral molecule DBT1 | | Descriptor: | 11-oxo-N-[2-(4-sulfamoylphenyl)ethyl]-10,11-dihydrodibenzo[b,f][1,4]thiazepine-8-carboxamide, Capsid protein | | Authors: | Schlicksup, C, Laughlin, P, Dunkelbarger, S, Wang, J.C, Zlotnick, A. | | Deposit date: | 2020-04-03 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Local Stabilization of Subunit-Subunit Contacts Causes Global Destabilization of Hepatitis B Virus Capsids.
Acs Chem.Biol., 15, 2020
|
|
5HQ3
 
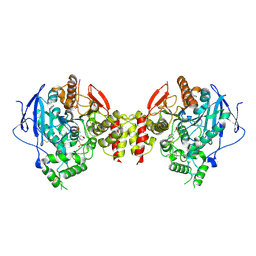 | | Stable, high-expression variant of human acetylcholinesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetylcholinesterase, O-ETHYLMETHYLPHOSPHONIC ACID ESTER GROUP | | Authors: | Goldenzweig, A, Goldsmith, M, Hill, S.E, Gertman, O, Laurino, P, Ashani, Y, Dym, O, Albeck, S, Unger, T, Prilusky, J, Lieberman, R.L, Aharoni, A, Silman, I, Sussman, J.L, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2016-01-21 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Automated Structure- and Sequence-Based Design of Proteins for High Bacterial Expression and Stability.
Mol.Cell, 63, 2016
|
|
4BRH
 
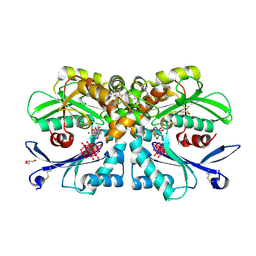 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG AND THIAMINE PHOSPHOVANADATE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DECAVANADATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic snapshots along the reaction pathway of nucleoside triphosphate diphosphohydrolases.
Structure, 21, 2013
|
|
3VC0
 
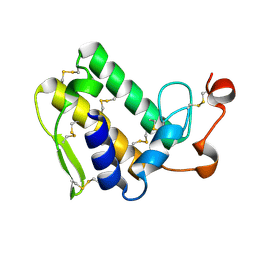 | | Crystal structure of Taipoxin beta subunit isoform 1 | | Descriptor: | Phospholipase A2 homolog, taipoxin beta chain | | Authors: | Cendron, L, Micetic, I, Polverino de Laureto, P, Beltramini, M, Paoli, M. | | Deposit date: | 2012-01-03 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of trimeric phospholipase A(2) neurotoxin from the Australian taipan snake venom.
Febs J., 279, 2012
|
|
7LOO
 
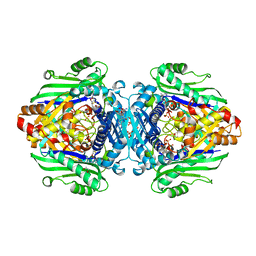 | | S-adenosyl methionine transferase cocrystallized with ATP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Jackson, C.J, Tan, L.L, Laurino, P. | | Deposit date: | 2021-02-10 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
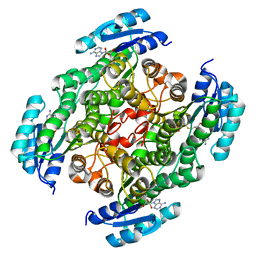
7XQM
 
 | |
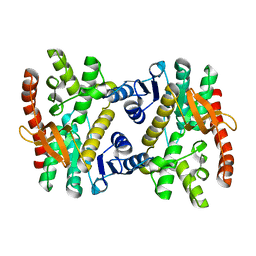
7XQN
 
 | |
4BRG
 
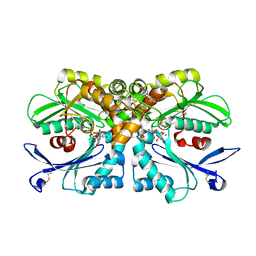 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG GMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BRF
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with a distorted orthomolybdate ion and AMP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
4BR7
 
 | | Legionella pneumophila NTPDase1 crystal form I, open, AMPNP complex | | Descriptor: | 5'-O-[(R)-hydroxy(phosphonoamino)phosphoryl]adenosine, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
