5XGN
 
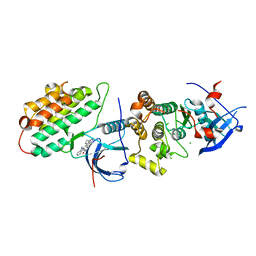 | | Crystal structure of EGFR 696-1022 T790M/C797S in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5XGM
 
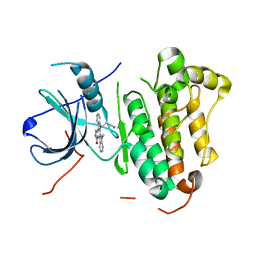 | | Crystal structure of EGFR 696-1022 T790M in complex with Go6976 | | Descriptor: | 12-(2-Cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole, Epidermal growth factor receptor | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2017-04-14 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.952 Å) | | Cite: | Structural pharmacological studies on EGFR T790M/C797S.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
5DH3
 
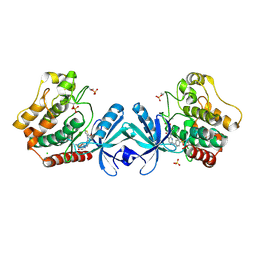 | | Crystal structure of MST2 in complex with XMU-MP-1 | | Descriptor: | 4-[(5,10-dimethyl-6-oxo-6,10-dihydro-5H-pyrimido[5,4-b]thieno[3,2-e][1,4]diazepin-2-yl)amino]benzenesulfonamide, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2015-08-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.468 Å) | | Cite: | Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration
Sci Transl Med, 8, 2016
|
|
5HU9
 
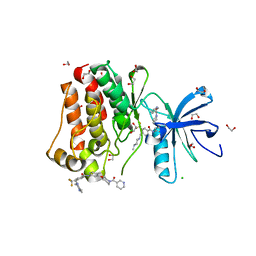 | | Crystal structure of ABL1 in complex with CHMFL-074 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylpiperazin-1-yl)methyl]-N-(4-methyl-3-{[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]oxy}phenyl)-3-(trifluoromethyl)benzamide, CHLORIDE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Discovery and characterization of a novel potent type II native and mutant BCR-ABL inhibitor (CHMFL-074) for Chronic Myeloid Leukemia (CML)
Oncotarget, 7, 2016
|
|
3JAU
 
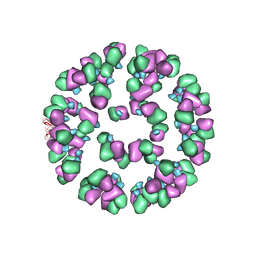 | | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | | Descriptor: | Capsid protein VP1, Heavy chain of Fab fragment variable region of antibody D5, Light chain of Fab fragment variable region of antibody D5 | | Authors: | Fan, C, Ye, X.H, Ku, Z.Q, Zuo, T, Kong, L.L, Zhang, C, Shi, J.P, Liu, Q.W, Chen, T, Zhang, Y.Y, Jiang, W, Zhang, L.Q, Huang, Z, Cong, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody
Plos Pathog., 12, 2016
|
|
7YAZ
 
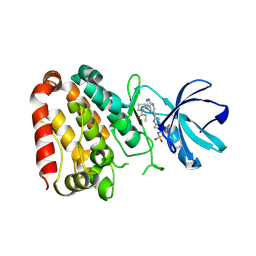 | | Crystal structure of ZAK in complex with compound YH-186 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-[3-[(3~{S})-1-propanoylpyrrolidin-3-yl]oxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl]-1,2,3-triazol-1-yl]phenyl]-3-phenyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
7YAW
 
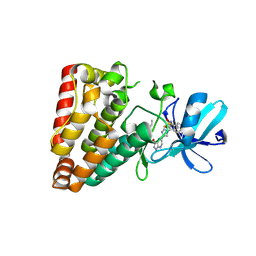 | | Crystal structure of ZAK in complex with compound YH-180 | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[3-[[5-[1-[2,6-bis(fluoranyl)-3-[(3-phenylphenyl)sulfonylamino]phenyl]-1,2,3-triazol-4-yl]-1~{H}-pyrazolo[3,4-b]pyridin-3-yl]oxy]propyl]propanamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2022-06-28 | | Release date: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational Design of Covalent Kinase Inhibitors by an Integrated Computational Workflow (Kin-Cov).
J.Med.Chem., 66, 2023
|
|
6JUU
 
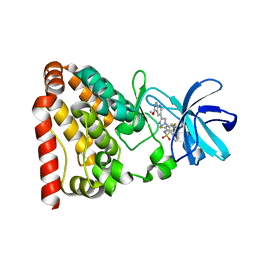 | | Crystal structure of ZAK in complex with compound 6r | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]naphthalene-1-sulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JUT
 
 | | Crystal structure of ZAK in complex with compound 6k | | Descriptor: | Mitogen-activated protein kinase kinase kinase MLT, ~{N}-[2,4-bis(fluoranyl)-3-[4-(3-methoxy-1~{H}-pyrazolo[3,4-b]pyridin-5-yl)-1,2,3-triazol-1-yl]phenyl]-3-bromanyl-benzenesulfonamide | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-alpha Motif Kinase (ZAK) Inhibitors.
J.Med.Chem., 63, 2020
|
|
6M3G
 
 | | Crystal structure of human HPF1 | | Descriptor: | Histone PARylation factor 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3I
 
 | | Crystal structure of HPF1/PARP1 complex | | Descriptor: | BENZAMIDE, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3H
 
 | | Crystal structure of mouse HPF1 | | Descriptor: | Histone PARylation factor 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
5TSL
 
 | |
5TSK
 
 | |
7WUQ
 
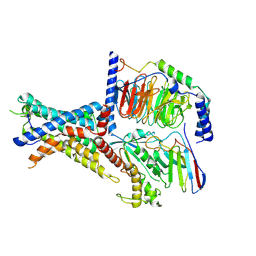 | | Tethered peptide activation mechanism of adhesion GPCRs ADGRG2 and ADGRG4 | | Descriptor: | Adhesion G-protein coupled receptor G2,mCherry, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Q.T, Guo, S.C, Xiao, P, Sun, J.P, Yu, X, Gou, L, Kong, L.L, Zhang, L. | | Deposit date: | 2022-02-09 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4.
Nature, 604, 2022
|
|
