8U2F
 
 | |
8XJU
 
 | |
8XJZ
 
 | | Cryo-EM structure of colibactin assembly line polyketide synthase ClbI KS-AT didomain crosslinked with its precursor module, ClbH | | Descriptor: | Peptide synthetase, Polyketide synthase, polypeptide from ClbH | | Authors: | Kim, M, Kim, J, Kang, J.Y. | | Deposit date: | 2023-12-22 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural study on human microbiome-derived polyketide synthases that assemble genotoxic colibactin.
Structure, 33, 2025
|
|
8XJY
 
 | |
9IRZ
 
 | | Crystal structure of YhaJ DNA-binding domain | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Probable HTH-type transcriptional regulator YhaJ | | Authors: | Kim, M, Kang, R, Ryu, S.E. | | Deposit date: | 2024-07-16 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | High-affinity promotor binding of YhaJ mediates a low signal leakage for effective DNT detection.
Front Microbiol, 15, 2024
|
|
4FEC
 
 | | Crystal Structure of Htt36Q3H | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FEB
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C2(Beta) | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, SODIUM ION, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FED
 
 | | Crystal Structure of Htt36Q3H | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-30 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
4FE8
 
 | | Crystal Structure of Htt36Q3H-EX1-X1-C1(Alpha) | | Descriptor: | Maltose-binding periplasmic protein,Huntingtin, ZINC ION | | Authors: | Kim, M. | | Deposit date: | 2012-05-29 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Beta conformation of polyglutamine track revealed by a crystal structure of Huntingtin N-terminal region with insertion of three histidine residues.
Prion, 7, 2013
|
|
8H5A
 
 | |
8H58
 
 | | Crystal structure of YhaJ effector binding domain | | Descriptor: | HTH-type transcriptional regulator YhaJ, SODIUM ION | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Structural basis of transcription factor YhaJ for DNT detection.
Iscience, 26, 2023
|
|
1IVW
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Late intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVX
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Holo form generated by biogenesis in crystal. | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVU
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Initial intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
1IVV
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis: Early intermediate in topaquinone biogenesis | | Descriptor: | COPPER (II) ION, amine oxidase | | Authors: | Kim, M, Okajima, T, Kishishita, S, Yoshimura, M, Kawamori, A, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2002-03-29 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray snapshots of quinone cofactor biogenesis in bacterial copper amine oxidase.
Nat.Struct.Biol., 9, 2002
|
|
7XC0
 
 | | Crystal structure of Human RPTPH | | Descriptor: | PHOSPHATE ION, Receptor-type tyrosine-protein phosphatase H | | Authors: | Kim, M, Ryu, S.E. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of the catalytic domain of human RPTPH.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
3GWL
 
 | |
3GWN
 
 | |
3P0K
 
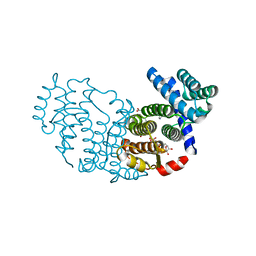 | | Structure of Baculovirus Sulfhydryl Oxidase Ac92 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2010-09-29 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure of a baculovirus sulfhydryl oxidase, a highly divergent member of the erv flavoenzyme family.
J.Virol., 85, 2011
|
|
3QZY
 
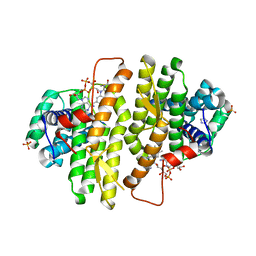 | | Structure of Baculovirus Sulfhydryl Oxidase Ac92 | | Descriptor: | Baculovirus sulfhydryl oxidase Ac92, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2011-03-07 | | Release date: | 2012-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of a baculovirus sulfhydryl oxidase, a highly divergent member of the erv flavoenzyme family.
J.Virol., 85, 2011
|
|
3TD7
 
 | | Crysal structure of the mimivirus sulfhydryl oxidase R596 | | Descriptor: | FAD-linked sulfhydryl oxidase R596, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hakim, M, Fass, D. | | Deposit date: | 2011-08-10 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Exploring ORFan domains in giant viruses: structure of mimivirus sulfhydryl oxidase R596.
Plos One, 7, 2012
|
|
8JCT
 
 | |
8XBL
 
 | |
8XJT
 
 | |
5Z7Q
 
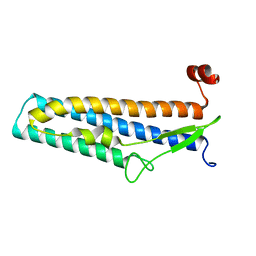 | | Crystal structure of Bacillus cereus flagellin | | Descriptor: | Flagellin | | Authors: | Kim, M, Hong, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Bacillus cereus flagellin and structure-guided fusion-protein designs
Sci Rep, 8, 2018
|
|
