7TVW
 
 | | Crystal structure of Arabidopsis thaliana DLK2 | | Descriptor: | Alpha/beta-Hydrolases superfamily protein | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Arabidopsis DWARF14-LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture.
Plant Direct, 6, 2022
|
|
6AZB
 
 | |
6AZD
 
 | |
6AZC
 
 | |
6ATX
 
 | |
6AVY
 
 | |
6AVW
 
 | |
6AVX
 
 | |
6AVV
 
 | |
3U0V
 
 | | Crystal Structure Analysis of human LYPLAL1 | | Descriptor: | Lysophospholipase-like protein 1 | | Authors: | Burger, M, Zimmermann, T.J, Kondoh, Y, Stege, P, Watanabe, N, Osada, H, Waldmann, H, Vetter, I.R. | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases
J.Lipid Res., 53, 2012
|
|
7UOC
 
 | | Crystal structure of Orobanche minor KAI2d4 | | Descriptor: | CHLORIDE ION, KAI2d4 | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Divergent Clade KAI2 Protein in the Root Parasitic Plant Orobanche minor Is a Highly Sensitive Strigolactone Receptor and Is Involved in the Perception of Sesquiterpene Lactones.
Plant Cell.Physiol., 64, 2023
|
|
6EZT
 
 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase D437A inactive mutant from Vibrio harveyi | | Descriptor: | Beta-N-acetylglucosaminidase Nag2, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
6EZR
 
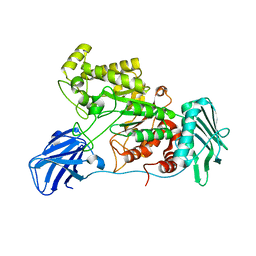 | |
6EZS
 
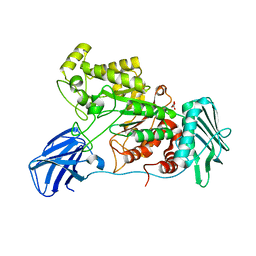 | | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi in complex with N-acetylglucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylglucosaminidase Nag2, MALONATE ION | | Authors: | Porfetye, A.T, Meekrathok, P, Burger, M, Vetter, I.R, Suginta, W. | | Deposit date: | 2017-11-16 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GH20 Exo beta-N-Acetylglucosaminidase from Vibrio harveyi
To Be Published
|
|
1DXM
 
 | | Reduced form of the H protein from glycine decarboxylase complex | | Descriptor: | DIHYDROLIPOIC ACID, H PROTEIN | | Authors: | Faure, M, Cohen-Addad, C, Neuburger, M, Douce, R. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
1HPC
 
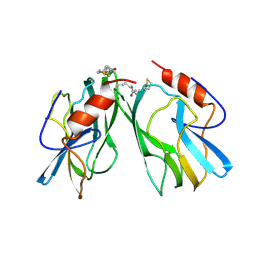 | | REFINED STRUCTURES AT 2 ANGSTROMS AND 2.2 ANGSTROMS OF THE TWO FORMS OF THE H-PROTEIN, A LIPOAMIDE-CONTAINING PROTEIN OF THE GLYCINE DECARBOXYLASE | | Descriptor: | 5-[(3S)-1,2-dithiolan-3-yl]pentanoic acid, H PROTEIN OF THE GLYCINE CLEAVAGE SYSTEM, LIPOIC ACID | | Authors: | Pares, S, Cohen-Addad, C, Sieker, L, Neuburger, M, Douce, R. | | Deposit date: | 1994-02-17 | | Release date: | 1995-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structures at 2 and 2.2 A resolution of two forms of the H-protein, a lipoamide-containing protein of the glycine decarboxylase complex.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1DXL
 
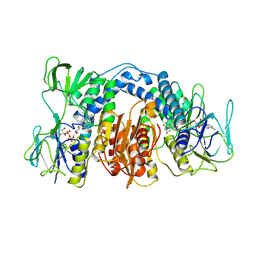 | | Dihydrolipoamide dehydrogenase of glycine decarboxylase from Pisum Sativum | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Faure, M, Cohen-Addad, C, Bourguignon, J, Macherel, D, Neuburger, M, Douce, R. | | Deposit date: | 2000-01-10 | | Release date: | 2000-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Interaction between the Lipoamide-Containing H-Protein and the Lipoamide Dehydrogenase (L-Protein) of the Glycine Decarboxylase Multienzyme System. 2. Crystal Structure of H- and L-Proteins
Eur.J.Biochem., 267, 2000
|
|
5DWR
 
 | | Identification of N-(4-((1R,3S,5S)-3-amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1,2 and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies | | Descriptor: | N-{4-[(1R,3S,5S)-3-amino-5-methylcyclohexyl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies.
J.Med.Chem., 58, 2015
|
|
5IIS
 
 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | Descriptor: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Bussiere, D, Burger, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-06 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4QAM
 
 | | Crystal Structure of the RPGR RCC1-like domain in complex with the RPGR-interacting domain of RPGRIP1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, X-linked retinitis pigmentosa GTPase regulator, ... | | Authors: | Remans, K, Buerger, M, Vetter, I.R, Wittinghofer, A. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | C2 domains as protein-protein interaction modules in the ciliary transition zone.
Cell Rep, 8, 2014
|
|
3SD5
 
 | | Crystal Structure of PI3K gamma with 5-(2,4-dimorpholinopyrimidin-6-yl)-4-(trifluoromethyl)pyridin-2-amine | | Descriptor: | 5-[2,6-di(morpholin-4-yl)pyrimidin-4-yl]-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2011-06-08 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification and Characterization of NVP-BKM120, an Orally Available Pan-Class I PI3-Kinase Inhibitor.
Mol.Cancer Ther., 11, 2012
|
|
4KZ0
 
 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, methyl 2-(acetylamino)-1,3-benzothiazole-6-carboxylate | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KZC
 
 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1HTP
 
 | |
4TN4
 
 | | Crystal structure of ternary complex of Plasmodium vivax SHMT with glycine and a novel pyrazolopyran 33G: (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4S)-6-amino-4-(5-cyano-3'-fluorobiphenyl-3-yl)-4-cyclobutyl-3-methyl-2,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Witschel, M.C. | | Deposit date: | 2014-06-03 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT): Cocrystal Structures of Pyrazolopyrans with Potent Blood- and Liver-Stage Activities.
J.Med.Chem., 58, 2015
|
|
