6ZP2
 
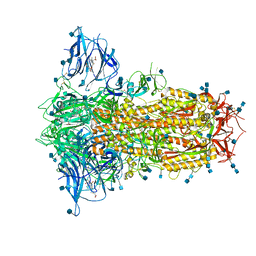 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP0
 
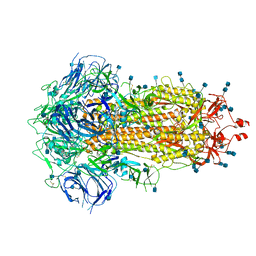 | | Structure of SARS-CoV-2 Spike Protein Trimer (single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZP1
 
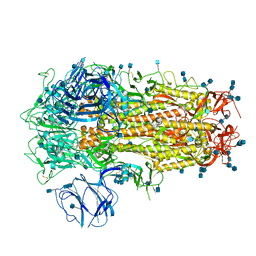 | | Structure of SARS-CoV-2 Spike Protein Trimer (K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOX
 
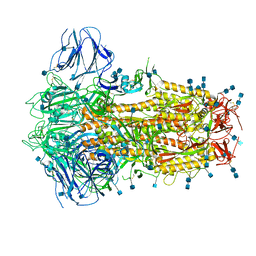 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x2 disulphide-bond mutant, G413C, V987C, single Arg S1/S2 cleavage site) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOY
 
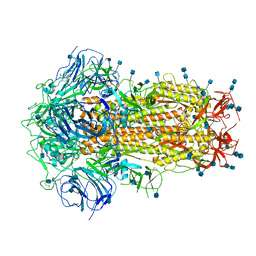 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Closed State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZOZ
 
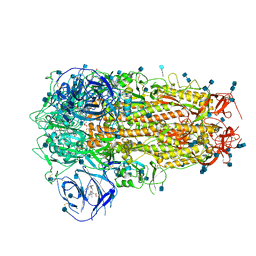 | | Structure of Disulphide-stabilized SARS-CoV-2 Spike Protein Trimer (x1 disulphide-bond mutant, S383C, D985C, K986P, V987P, single Arg S1/S2 cleavage site) in Locked State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Xiong, X, Qu, K, Scheres, S.H.W, Briggs, J.A.G. | | Deposit date: | 2020-07-08 | | Release date: | 2020-07-22 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A thermostable, closed SARS-CoV-2 spike protein trimer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1O9A
 
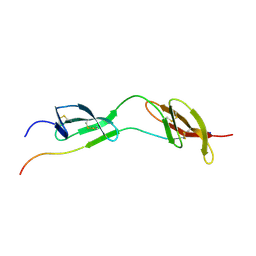 | | Solution structure of the complex of 1F12F1 from fibronectin with B3 from FnBB from S. dysgalactiae | | Descriptor: | FIBRONECTIN, FIBRONECTIN BINDING PROTEIN | | Authors: | Schwarz-Linek, U, Werner, J.M, Pickford, A.R, Pilka, E.S, Gurusiddappa, S, Briggs, J.A.G, Hook, M, Campbell, I.D, Potts, J.R. | | Deposit date: | 2002-12-11 | | Release date: | 2003-05-08 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper.
Nature, 423, 2003
|
|
5L93
 
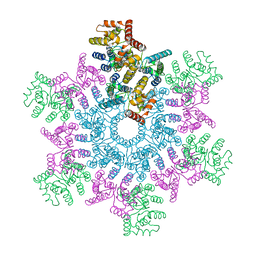 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | Descriptor: | Capsid protein p24 | | Authors: | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
4UFC
 
 | | Crystal structure of the GH95 enzyme BACOVA_03438 | | Descriptor: | CACODYLATE ION, CALCIUM ION, GH95, ... | | Authors: | Rogowski, A, Briggs, J.A, Mortimer, J.C, Tryfona, T, Terrapon, N, Lowe, E.C, Basle, A, Morland, C, Day, A.M, Zheng, H, Rogers, T.E, Thompson, P, Hawkins, A.R, Yadav, M.P, Henrissat, B, Martens, E.C, Dupree, P, Gilbert, H.J, Bolam, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-07-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Glycan Complexity Dictates Microbial Resource Allocation in the Large Intestine.
Nat.Commun., 6, 2015
|
|
7Z5C
 
 | | Chimera of AP2 with FCHO2 linker domain as a fusion on Cmu2 subunit | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Kane Dickson, V, Qu, K, Owen, D.J, Briggs, J.A, Zaccai, N.R. | | Deposit date: | 2022-03-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
5MD9
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=6 | | Descriptor: | Capsid protein p24 C-terminal domain, Capsid protein p24 N-terminal domain | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD7
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=-12 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD3
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=12 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDE
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=23, twist=0 | | Descriptor: | Capsid protein p24 C-terminal domain, Capsid protein p24 N-terminal domain | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MCZ
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=-1, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDB
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=-6 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD8
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=17, twist=12 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MCY
 
 | | The structure of the mature HIV-1 CA pentamer in intact virus particles | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD6
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=-6 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD5
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=11, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDD
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=23, twist=6 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD1
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=5, twist=0 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MCX
 
 | | The structure of the mature HIV-1 CA hexamer in intact virus particles | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MD0
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=5, twist=6 | | Descriptor: | Capsid protein p24 | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
5MDF
 
 | | The structure of the mature HIV-1 CA hexameric lattice with curvature parameters: tilt=23, twist=-6 | | Descriptor: | Gag protein | | Authors: | Mattei, S, Glass, B, Hagen, W.J.H, Kraeusslich, H.-G, Briggs, J.A.G. | | Deposit date: | 2016-11-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Science, 354, 2016
|
|
