4CMH
 
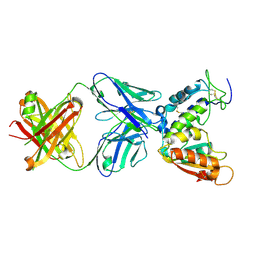 | | Crystal structure of CD38 with a novel CD38-targeting antibody SAR650984 | | Descriptor: | ADP-RIBOSYL CYCLASE 1, HEAVY CHAIN OF SAR650984-FAB FRAGMENT, LIGHT CHAIN OF SAR650984-FAB FRAGMENT | | Authors: | Deckert, J, Wetzel, M.C, Park, P.U, Bartle, L.M, Skaletskaya, A, Goldmacher, V, Vallee, F, ZhouLiu, Q, Ferrari, P, Pouzieux, S, Lahoute, C, Dumontet, C, Plesa, A, Chiron, M, Lejeune, P, Chittenden, T, Blanc, V. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | SAR650984, a novel humanized CD38-targeting antibody, demonstrates potent antitumor activity in models of multiple myeloma and other CD38+ hematologic malignancies.
Clin. Cancer Res., 20, 2014
|
|
1LRY
 
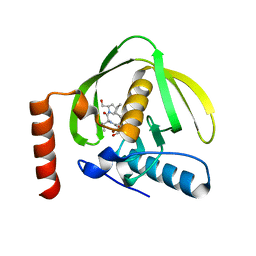 | | Crystal Structure of P. aeruginosa Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE deformylase, ZINC ION | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1LRU
 
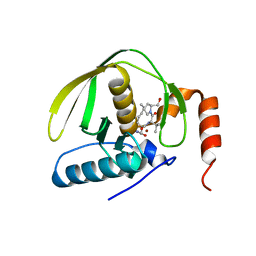 | | Crystal Structure of E.coli Peptide Deformylase Complexed with Antibiotic Actinonin | | Descriptor: | ACTINONIN, PEPTIDE DEFORMYLASE, SULFATE ION, ... | | Authors: | Guilloteau, J.-P, Mathieu, M, Giglione, C, Blanc, V, Dupuy, A, Chevrier, M, Gil, P, Famechon, A, Meinnel, T, Mikol, V. | | Deposit date: | 2002-05-16 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1LQW
 
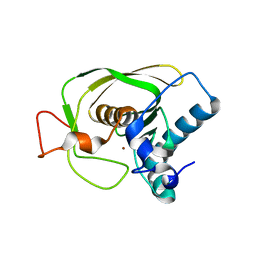 | | Crystal Structure of S.aureus Peptide Deformylase | | Descriptor: | PEPTIDE DEFORMYLASE PDF1, ZINC ION | | Authors: | Mikol, V. | | Deposit date: | 2002-05-14 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
1LQY
 
 | |
3ELM
 
 | | Crystal Structure of MMP-13 Complexed with Inhibitor 24f | | Descriptor: | (2R)-({[5-(4-ethoxyphenyl)thiophen-2-yl]sulfonyl}amino){1-[(1-methylethoxy)carbonyl]piperidin-4-yl}ethanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Kulathila, R, Monovich, L, Koehn, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent, selective, and orally active carboxylic acid based inhibitors of matrix metalloproteinase-13
J.Med.Chem., 52, 2009
|
|
