6JT4
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4S,6S)-2-amino-4-methyl-6-(trifluoromethyl)-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-5-(fluoromethoxy)pyrazine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Anan, K, Iso, Y, Oguma, T, Nakahara, K, Suzuki, S, Yamamoto, T, Matsuoka, E, Ito, H, Sakaguchi, G, Ando, S, Morimoto, K, Kanegawa, N, Kido, Y, Kawachi, T, Fukushima, T, Teisman, A, Urmaliya, V, Dhuyvetter, D, Borghys, H, Austin, N, Bergh, A.V.D, Verboven, P, Bischoff, F, Gijsen, H.J.M, Yamano, Y, Kusakabe, K.I. | | Deposit date: | 2019-04-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trifluoromethyl Dihydrothiazine-Based beta-Secretase (BACE1) Inhibitors with Robust Central beta-Amyloid Reduction and Minimal Covalent Binding Burden.
Chemmedchem, 14, 2019
|
|
9FMR
 
 | |
2W2H
 
 | |
2VB0
 
 | | Crystal structure of coxsackievirus B3 proteinase 3C | | Descriptor: | CHLORIDE ION, POLYPROTEIN 3BCD | | Authors: | Anand, K, Mesters, J.R, Goerlach, R, Zell, R, Hilgenfeld, R. | | Deposit date: | 2007-09-05 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Coxsackie Virus B3 Proteinase 3C
To be Published
|
|
4A6F
 
 | | Crystal structure of Slm1-PH domain in complex with Phosphoserine | | Descriptor: | PHOSPHATE ION, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE-BINDING PROTEIN SLM1, PHOSPHOSERINE | | Authors: | Anand, K, Maeda, K, Gavin, A.C. | | Deposit date: | 2011-11-02 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Analyses of Slm1-Ph Domain Demonstrate Ligand Binding in the Non-Canonical Site
Plos One, 7, 2012
|
|
4A6K
 
 | | Crystal structure of Slm1-PH domain in complex with D-myo-Inositol-4- phosphate | | Descriptor: | D-MYO-INOSITOL-4-PHOSPHATE, PHOSPHATE ION, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE-BINDING PROTEIN SLM1 | | Authors: | Anand, K, Maeda, K, Gavin, A.C. | | Deposit date: | 2011-11-04 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Analyses of Slm1-Ph Domain Demonstrate Ligand Binding in the Non-Canonical Site
Plos One, 7, 2012
|
|
4A6H
 
 | | Crystal structure of Slm1-PH domain in complex with Inositol-4- phosphate | | Descriptor: | D-MYO-INOSITOL-4-PHOSPHATE, PHOSPHATE ION, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE-BINDING PROTEIN SLM1 | | Authors: | Anand, K, Maeda, K, Gavin, A.C. | | Deposit date: | 2011-11-03 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structural Analyses of Slm1-Ph Domain Demonstrate Ligand Binding in the Non-Canonical Site
Plos One, 7, 2012
|
|
4A5K
 
 | |
2PK2
 
 | | Cyclin box structure of the P-TEFb subunit Cyclin T1 derived from a fusion complex with EIAV Tat | | Descriptor: | Cyclin-T1, Protein Tat | | Authors: | Anand, K, Schulte, A, Fujinaga, K, Scheffzek, K, Geyer, M. | | Deposit date: | 2007-04-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Cyclin Box Structure of the P-TEFb Subunit Cyclin T1 Derived from a Fusion Complex with EIAV Tat.
J.Mol.Biol., 370, 2007
|
|
2WU8
 
 | |
3IPV
 
 | | Crystal structure of Spatholobus parviflorus seed lectin | | Descriptor: | CALCIUM ION, Lectin alpha chain, Lectin beta chain, ... | | Authors: | Geethanandan, K, Bharath, S.R, Abhilash, J, Sadasivan, C, Haridas, M. | | Deposit date: | 2009-08-18 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | X-ray structure of a galactose-specific lectin from Spatholobous parviflorous
Int.J.Biol.Macromol., 49, 2011
|
|
2X1V
 
 | |
1P9U
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PHQ-VNSTLQ-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION, ... | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
1P9S
 
 | | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replicase polyprotein 1ab | | Authors: | Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2003-05-12 | | Release date: | 2003-05-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Coronavirus Main Proteinase (3CLpro) Structure: Basis for Design of anti-SARS Drugs
Science, 300, 2003
|
|
3DGV
 
 | | Crystal structure of thrombin activatable fibrinolysis inhibitor (TAFI) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Anand, K, Pallares, I, Valnickova, Z, Christensen, T, Schreuder, H, Enghild, J. | | Deposit date: | 2008-06-16 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of thrombin-activable fibrinolysis inhibitor (TAFI) provides the structural basis for its intrinsic activity and the short half-life of TAFIa.
J.Biol.Chem., 283, 2008
|
|
1LVO
 
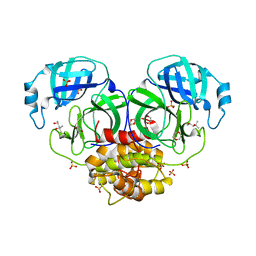 | | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1,4-DIETHYLENE DIOXIDE, Replicase, ... | | Authors: | Anand, K, Palm, G.J, Mesters, J.R, Siddell, S.G, Ziebuhr, J, Hilgenfeld, R. | | Deposit date: | 2002-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra alpha-helical domain.
EMBO J., 21, 2002
|
|
8PYR
 
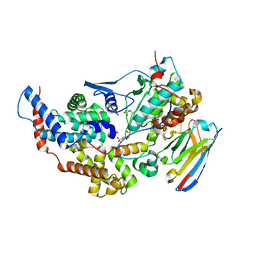 | | Crystal structure of the dual T-loop phosphorylated Cdk7/CycH/Mat1 complex | | Descriptor: | 1,2-ETHANEDIOL, CDK-activating kinase assembly factor MAT1, Cyclin-H, ... | | Authors: | Anand, K, Duster, R, Geyer, M. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis of Cdk7 activation by dual T-loop phosphorylation.
Biorxiv, 2024
|
|
7BAI
 
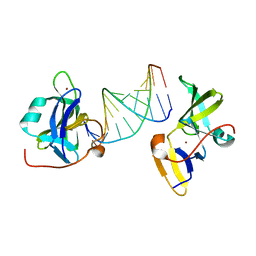 | |
7BAH
 
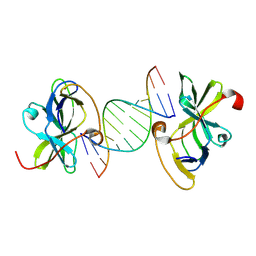 | | Structure of RIG-I CTD bound to OH-RNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, RNA (5'-R(*GP*AP*CP*GP*CP*UP*AP*GP*CP*GP*UP*C)-3'), ZINC ION | | Authors: | Anand, K, Hagelueken, G, Fusshoeller, D, Geyer, M. | | Deposit date: | 2020-12-15 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A conserved isoleucine in the RNA sensor RIG-I controls immune tolerance to mitochondrial RNA
To Be Published
|
|
7NXK
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with the inhibitor BSJ-01-175 | | Descriptor: | (E)-N-[4-[(1R,3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]cyclohexyl]oxyphenyl]-4-(dimethylamino)but-2-enamide, Cyclin-K, Cyclin-dependent kinase 12 | | Authors: | Anand, K, Dust, S, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
7NXJ
 
 | | Crystal structure of human Cdk13/Cyclin K in complex with the inhibitor THZ531 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 13, N-[4-[(3R)-3-[[5-chloranyl-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]piperidin-1-yl]carbonylphenyl]-4-(dimethylamino)butanamide | | Authors: | Anand, K, Greifenberg, A.K, Kaltheuner, I.H, Geyer, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Eur.J.Med.Chem., 221, 2021
|
|
8P81
 
 | | Crystal structure of human Cdk12/Cyclin K in complex with inhibitor SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, ~{N}-[[5,6-bis(chloranyl)-1~{H}-benzimidazol-2-yl]methyl]-9-(1-methylpyrazol-4-yl)-2-morpholin-4-yl-purin-6-amine | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2023-05-31 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The reversible inhibitor SR-4835 binds Cdk12/cyclin K in a noncanonical G-loop conformation.
J.Biol.Chem., 300, 2023
|
|
2A7T
 
 | | Crystal Structure of a novel neurotoxin from Buthus tamalus at 2.2A resolution. | | Descriptor: | Neurotoxin | | Authors: | Ethayathulla, A.S, Sharma, M, Saravanan, K, Sharma, S, Kaur, P, Yadav, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2005-07-06 | | Release date: | 2005-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a highly acidic neurotoxin from scorpion Buthus tamulus at 2.2A resolution reveals novel structural features.
J.Struct.Biol., 155, 2006
|
|
3ZYD
 
 | | Crystal structure of 3C protease of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE, GLYCEROL | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Peptidic Ab-Nonsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZYE
 
 | | Crystal structure of 3C protease mutant (T68A) of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
