4UXE
 
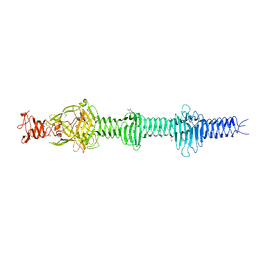 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 selenomethionine crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-19 | | Last modified: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4UXG
 
 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, R32 native crystal | | Descriptor: | LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
4UXF
 
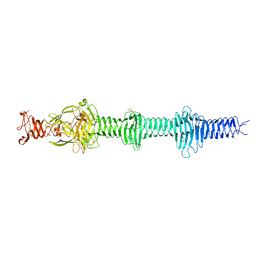 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, P21 native crystal | | Descriptor: | GLYCEROL, LARGE TAIL FIBER PROTEIN P34 | | Authors: | Granell, M, Alvira, S, Garcia-Doval, C, Singh, A.K, van Raaij, M.J. | | Deposit date: | 2014-08-22 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
5NXF
 
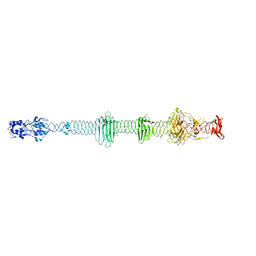 | | Crystal structure of the carboxy-terminal region of the bacteriophage T4 proximal long tail fibre protein gp34, residues 795 to 1289, at 1.9 Angstrom. | | Descriptor: | ACETATE ION, GLYCEROL, Long-tail fiber proximal subunit, ... | | Authors: | Namura, M, van Raaij, M.J, Kanamaru, S. | | Deposit date: | 2017-05-10 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Carboxy-Terminal Region of the Bacteriophage T4 Proximal Long Tail Fiber Protein Gp34.
Viruses, 9, 2017
|
|
5NXH
 
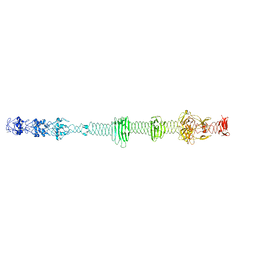 | |
7PIM
 
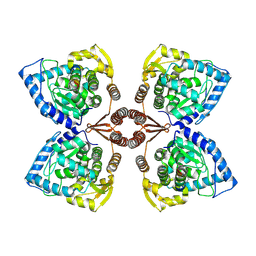 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | Descriptor: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6QIY
 
 | | CI-2, conformation 1 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6QIZ
 
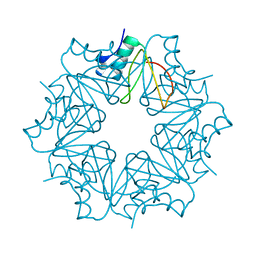 | | CI-2, conformation 2 | | Descriptor: | Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Romero, A, Ruiz, F.M. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Engineering protein assemblies with allosteric control via monomer fold-switching.
Nat Commun, 10, 2019
|
|
6ZVP
 
 | | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | | Descriptor: | FE (III) ION, L-DOPAMINE, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-27 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZZU
 
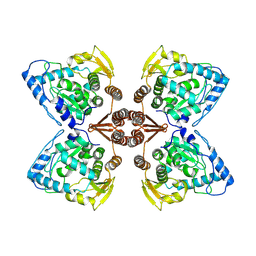 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-08-05 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7A2G
 
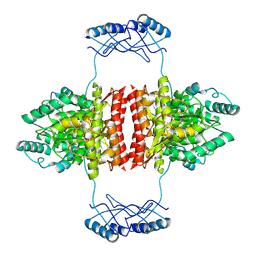 | | Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Flydal, M.I, Martinez, A, Valpuesta, J.M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZN2
 
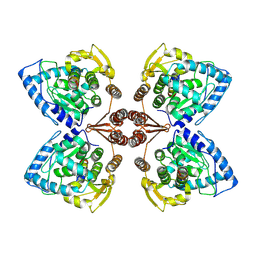 | | Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding. | | Descriptor: | FE (III) ION, L-DOPAMINE, SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-ALA-ALA, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
