1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
1P9Q
 
 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1QW2
 
 | | Crystal Structure of a Protein of Unknown Function TA1206 from Thermoplasma acidophilum | | Descriptor: | conserved hypothetical protein TA1206 | | Authors: | Savchenko, A, Evdokimova, E, Kudrytska, M, Edwards, A.E, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-30 | | Release date: | 2004-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hypothetical Protein "TA1206" from Thermoplasma acidophilum
To be Published
|
|
4W8K
 
 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
1K7K
 
 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2PYU
 
 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with IMP | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, Inosine Triphosphate Pyrophosphatase RdgB | | Authors: | Singer, A.U, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2Q16
 
 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with ITP | | Descriptor: | CALCIUM ION, HAM1 protein homolog, INOSINE 5'-TRIPHOSPHATE, ... | | Authors: | Singer, A.U, Lam, R, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1TF1
 
 | | Crystal Structure of the E. coli Glyoxylate Regulatory Protein Ligand Binding Domain | | Descriptor: | Negative regulator of allantoin and glyoxylate utilization operons | | Authors: | Walker, J.R, Skarina, T, Kudrytska, M, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-26 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical study of effector molecule recognition by the E.coli glyoxylate and allantoin utilization regulatory protein AllR.
J.Mol.Biol., 358, 2006
|
|
1TD5
 
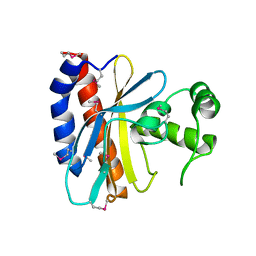 | | Crystal Structure of the Ligand Binding Domain of E. coli IclR. | | Descriptor: | Acetate operon repressor | | Authors: | Walker, J.R, Evdokimova, L, Zhang, R.-G, Bochkarev, A, Joachimiak, A, Arrowsmith, C, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analyses of the Ligand Binding Sites of the IclR family of transcriptional regulators
To be Published
|
|
3K67
 
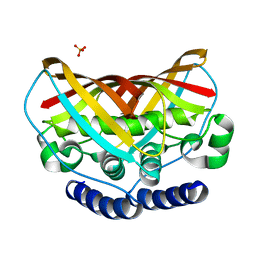 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
8U7F
 
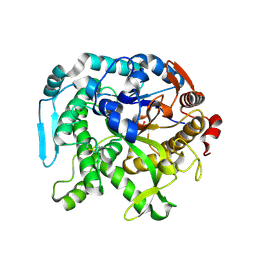 | | Crystal structure of CIB_12 beta-galactosidase from Cuniculiplasma divulgatum | | Descriptor: | CIB_12 Beta-galactosidase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yakunin, A, Golyshin, P, Savchenko, A. | | Deposit date: | 2023-09-15 | | Release date: | 2024-07-24 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Moderately thermostable GH1 beta-glucosidases from hyperacidophilic archaeon Cuniculiplasma divulgatum S5.
Fems Microbiol.Ecol., 100, 2024
|
|
3K3S
 
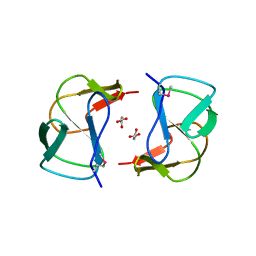 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
4KI3
 
 | | 1.70 Angstrom resolution crystal structure of outer-membrane lipoprotein carrier protein (lolA) from Yersinia pestis CO92 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-05-01 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.70 Angstrom resolution crystal structure of outer-membrane lipoprotein carrier protein (lolA) from Yersinia pestis CO92
To be Published
|
|
1TE2
 
 | | Putative Phosphatase Ynic from Escherichia coli K12 | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, 2-deoxyglucose-6-P phosphatase, CALCIUM ION | | Authors: | Kim, Y, Joachimiak, A, Evdokimova, E, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-24 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Putative Phosphatase Ynic from Escherichia coli K12
To be Published
|
|
1JQ3
 
 | | Crystal Structure of Spermidine Synthase in Complex with Transition State Analogue AdoDATO | | Descriptor: | S-ADENOSYL-1,8-DIAMINO-3-THIOOCTANE, Spermidine synthase | | Authors: | Korolev, S, Ikeguchi, Y, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Pegg, A.E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
1K4N
 
 | | Structural Genomics, Protein EC4020 | | Descriptor: | Protein EC4020 | | Authors: | Zhang, R.G, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-08 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
Proteins, 51, 2003
|
|
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
6HCD
 
 | | Structure of universal stress protein from Archaeoglobus fulgidus | | Descriptor: | ACETATE ION, CHLORIDE ION, UNIVERSAL STRESS PROTEIN, ... | | Authors: | Shumilin, I.A, Loch, J.I, Cymborowski, M, Xu, X, Edwards, A, Di Leo, R, Shabalin, I.G, Joachimiak, A, Savchenko, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2018-08-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the universal stress protein family.
Evol Appl, 6, 2013
|
|
9BIY
 
 | |
9BIZ
 
 | |
9BJ0
 
 | |
1SR8
 
 | | Structural Genomics, 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus | | Descriptor: | cobalamin biosynthesis protein (cbiD) | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of cobalamin biosynthesis protein (cbiD) from Archaeoglobus fulgidus
To be Published
|
|
1T07
 
 | | Crystal Structure of Conserved Protein of Unknown Function PA5148 from Pseudomonas aeruginosa | | Descriptor: | Hypothetical UPF0269 protein PA5148 | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-07 | | Release date: | 2004-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of PA5148 from Pseudomonas aeruginosa
To be Published
|
|
1T1J
 
 | | Crystal structure of genomics APC5043 | | Descriptor: | hypothetical protein | | Authors: | Dong, A, Xu, X, Liu, Y, Zhang, R, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein PA1492 from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
